Gene Page: CDK5RAP3
Summary ?
| GeneID | 80279 |
| Symbol | CDK5RAP3 |
| Synonyms | C53|HSF-27|IC53|LZAP|MST016|OK/SW-cl.114|PP1553 |
| Description | CDK5 regulatory subunit associated protein 3 |
| Reference | MIM:608202|HGNC:HGNC:18673|Ensembl:ENSG00000108465|HPRD:12190|Vega:OTTHUMG00000178682 |
| Gene type | protein-coding |
| Map location | 17q21.32 |
| Pascal p-value | 0.23 |
| Sherlock p-value | 0.345 |
| Fetal beta | 1.489 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Cerebellar Hemisphere Cerebellum Frontal Cortex BA9 Putamen basal ganglia Myers' cis & trans |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14007442 | 17 | 46048497 | CDK5RAP3 | 1.08E-7 | -0.006 | 2.35E-5 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16955618 | chr15 | 29937543 | CDK5RAP3 | 80279 | 0.16 | trans | ||
| rs8073326 | 17 | 46509708 | CDK5RAP3 | ENSG00000108465.10 | 2.70752E-7 | 0.01 | 464532 | gtex_brain_ba24 |
| rs7211636 | 17 | 46512688 | CDK5RAP3 | ENSG00000108465.10 | 1.26501E-7 | 0.01 | 467512 | gtex_brain_ba24 |
| rs4267363 | 17 | 46524294 | CDK5RAP3 | ENSG00000108465.10 | 1.24295E-7 | 0.01 | 479118 | gtex_brain_ba24 |
| rs6504256 | 17 | 46525183 | CDK5RAP3 | ENSG00000108465.10 | 1.21894E-7 | 0.01 | 480007 | gtex_brain_ba24 |
| rs4794678 | 17 | 46526303 | CDK5RAP3 | ENSG00000108465.10 | 5.90189E-8 | 0.01 | 481127 | gtex_brain_ba24 |
| rs1858162 | 17 | 46532294 | CDK5RAP3 | ENSG00000108465.10 | 1.19911E-7 | 0.01 | 487118 | gtex_brain_ba24 |
| rs11657639 | 17 | 46547464 | CDK5RAP3 | ENSG00000108465.10 | 6.80404E-7 | 0.01 | 502288 | gtex_brain_ba24 |
| rs117972490 | 17 | 46046649 | CDK5RAP3 | ENSG00000108465.10 | 4.01573E-6 | 0.04 | 1473 | gtex_brain_putamen_basal |
| rs61454280 | 17 | 46050125 | CDK5RAP3 | ENSG00000108465.10 | 4.0148E-6 | 0.04 | 4949 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
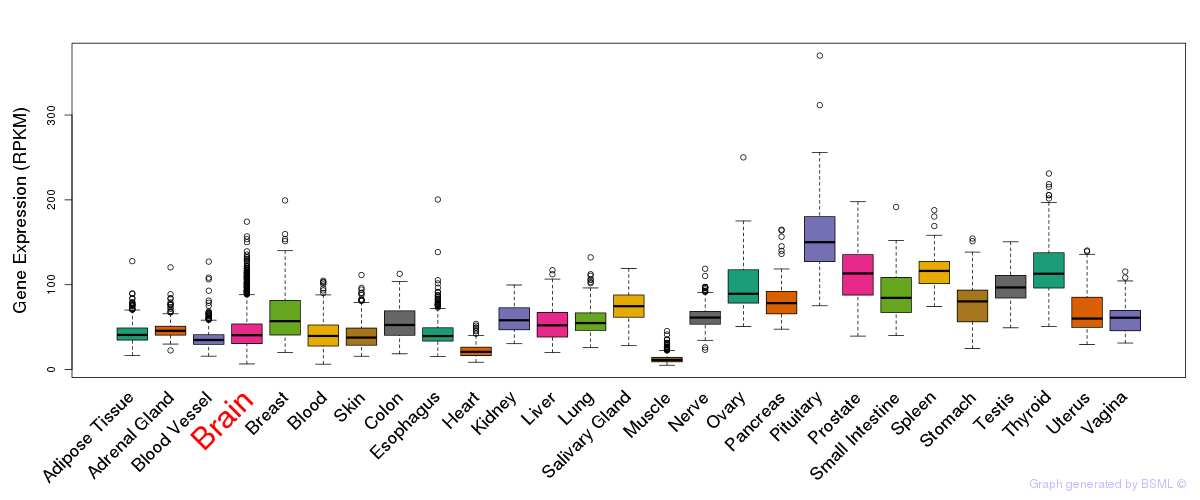
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0042808 | neuronal Cdc2-like kinase binding | NAS | neuron (GO term level: 7) | 10721722 |
| GO:0005515 | protein binding | IPI | 17043677 |17353931 |17785205 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007420 | brain development | NAS | Brain (GO term level: 7) | 10915792 |
| GO:0045664 | regulation of neuron differentiation | NAS | neuron, neurogenesis (GO term level: 9) | 10721722 |
| GO:0000079 | regulation of cyclin-dependent protein kinase activity | ISS | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005575 | cellular_component | ND | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ARL1 | ARFL1 | ADP-ribosylation factor-like 1 | Affinity Capture-MS | BioGRID | 17353931 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | - | HPRD | 10721722 |10915792 |
| CDK5R1 | CDK5P35 | CDK5R | MGC33831 | NCK5A | p23 | p25 | p35 | p35nck5a | cyclin-dependent kinase 5, regulatory subunit 1 (p35) | - | HPRD,BioGRID | 11882646 |
| CRMP1 | DPYSL1 | DRP-1 | DRP1 | collapsin response mediator protein 1 | Two-hybrid | BioGRID | 16169070 |
| DDRGK1 | C20orf116 | MGC2592 | dJ1187M17.3 | DDRGK domain containing 1 | Affinity Capture-MS | BioGRID | 17353931 |
| IGHA1 | FLJ14473 | FLJ35065 | FLJ35500 | FLJ36402 | FLJ39698 | FLJ40001 | FLJ41548 | FLJ41552 | FLJ41789 | FLJ43248 | FLJ43594 | FLJ44293 | FLJ46028 | FLJ46621 | FLJ46724 | FLJ46811 | FLJ46824 | IgA1 | MGC102857 | immunoglobulin heavy constant alpha 1 | Affinity Capture-MS | BioGRID | 17353931 |
| IGKC | HCAK1 | Km | MGC111575 | MGC62011 | MGC72072 | MGC88770 | MGC88771 | MGC88809 | immunoglobulin kappa constant | Affinity Capture-MS | BioGRID | 17353931 |
| KHK | - | ketohexokinase (fructokinase) | Affinity Capture-MS | BioGRID | 17353931 |
| KIAA0776 | RP3-393D12.1 | KIAA0776 | Affinity Capture-MS | BioGRID | 17353931 |
| SMNDC1 | SMNR | SPF30 | survival motor neuron domain containing 1 | Affinity Capture-MS | BioGRID | 17353931 |
| UFC1 | HSPC155 | ubiquitin-fold modifier conjugating enzyme 1 | Affinity Capture-MS | BioGRID | 17353931 |
| UFM1 | BM-002 | C13orf20 | bA131P10.1 | ubiquitin-fold modifier 1 | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| LAIHO COLORECTAL CANCER SERRATED DN | 86 | 47 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS DN | 145 | 93 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |