Gene Page: ACTN4
Summary ?
| GeneID | 81 |
| Symbol | ACTN4 |
| Synonyms | ACTININ-4|FSGS|FSGS1 |
| Description | actinin alpha 4 |
| Reference | MIM:604638|HGNC:HGNC:166|HPRD:05222| |
| Gene type | protein-coding |
| Map location | 19q13 |
| Pascal p-value | 0.548 |
| Fetal beta | -0.814 |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanNRC CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
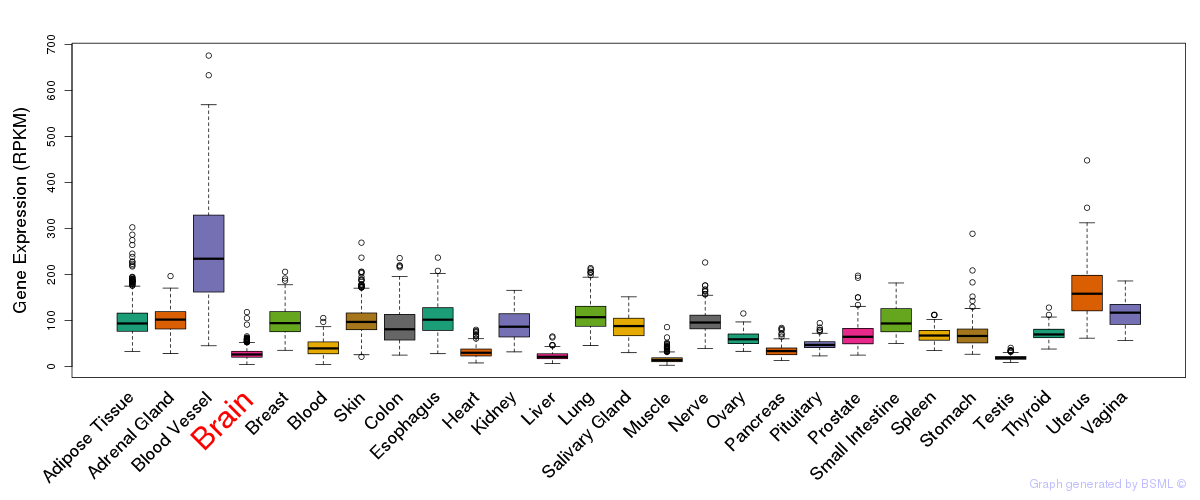
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTC1 | ACTC | CMD1R | CMH11 | actin, alpha, cardiac muscle 1 | - | HPRD | 12042308 |
| ATP6V1B1 | ATP6B1 | MGC32642 | RTA1B | VATB | VMA2 | VPP3 | ATPase, H+ transporting, lysosomal 56/58kDa, V1 subunit B1 | - | HPRD | 10915794 |
| CAMK2A | CAMKA | KIAA0968 | calcium/calmodulin-dependent protein kinase II alpha | CAMK2A (CaMKII-alpha) interacts with ACTN4 (alpha-Actinin 4). | BIND | 11160423 |
| CAMK2A | CAMKA | KIAA0968 | calcium/calmodulin-dependent protein kinase II alpha | Two-hybrid | BioGRID | 11160423 |
| CAMK2B | CAM2 | CAMK2 | CAMKB | MGC29528 | calcium/calmodulin-dependent protein kinase II beta | Two-hybrid | BioGRID | 11160423 |
| CAMK2B | CAM2 | CAMK2 | CAMKB | MGC29528 | calcium/calmodulin-dependent protein kinase II beta | CAMK2B (CaMKII-beta) interacts with ACTN4 (alpha-Actinin 4). | BIND | 11160423 |
| CAMK2G | CAMK | CAMK-II | CAMKG | FLJ16043 | MGC26678 | calcium/calmodulin-dependent protein kinase II gamma | - | HPRD | 11160423 |
| COL17A1 | BA16H23.2 | BP180 | BPAG2 | FLJ60881 | KIAA0204 | LAD-1 | collagen, type XVII, alpha 1 | - | HPRD,BioGRID | 11739652 |
| GSN | DKFZp313L0718 | gelsolin (amyloidosis, Finnish type) | - | HPRD,BioGRID | 11733011 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | - | HPRD | 2116421 |
| LRRC7 | DKFZp686I1147 | KIAA1365 | MGC144918 | leucine rich repeat containing 7 | - | HPRD | 11160423 |
| LRRC7 | DKFZp686I1147 | KIAA1365 | MGC144918 | leucine rich repeat containing 7 | LRRC7 (Densin-180) interacts with ACTN4 (alpha-Actinin 4). This interaction was modeled on a demonstrated interaction between rat LRCC7 and human ACTN4. | BIND | 11160423 |
| MAGI1 | AIP3 | BAIAP1 | BAP1 | MAGI-1 | TNRC19 | WWP3 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | - | HPRD,BioGRID | 12042308 |
| MYOZ1 | CS-2 | FATZ | MYOZ | myozenin 1 | Two-hybrid | BioGRID | 16189514 |
| MYOZ2 | C4orf5 | CS-1 | myozenin 2 | Two-hybrid | BioGRID | 16189514 |
| NEB | DKFZp686C1456 | FLJ11505 | FLJ36536 | FLJ39568 | FLJ39584 | NEB177D | NEM2 | nebulin | - | HPRD | 11882289 |
| NOS2 | HEP-NOS | INOS | NOS | NOS2A | nitric oxide synthase 2, inducible | - | HPRD | 12960352 |
| PDLIM1 | CLIM1 | CLP-36 | CLP36 | hCLIM1 | PDZ and LIM domain 1 | - | HPRD,BioGRID | 10753915 |
| PDLIM1 | CLIM1 | CLP-36 | CLP36 | hCLIM1 | PDZ and LIM domain 1 | CLP-36 interacts with alpha-actinin 4. | BIND | 10753915 |
| SLC9A3R2 | E3KARP | MGC104639 | NHE3RF2 | NHERF-2 | NHERF2 | OCTS2 | SIP-1 | SIP1 | TKA-1 | solute carrier family 9 (sodium/hydrogen exchanger), member 3 regulator 2 | - | HPRD,BioGRID | 11948184 |
| TRIM3 | BERP | FLJ16135 | HAC1 | RNF22 | RNF97 | tripartite motif-containing 3 | - | HPRD,BioGRID | 10673389 |
| USP6NL | KIAA0019 | RNTRE | TRE2NL | USP6 N-terminal like | - | HPRD,BioGRID | 15152255 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG FOCAL ADHESION | 201 | 138 | All SZGR 2.0 genes in this pathway |
| KEGG ADHERENS JUNCTION | 75 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG TIGHT JUNCTION | 134 | 86 | All SZGR 2.0 genes in this pathway |
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG REGULATION OF ACTIN CYTOSKELETON | 216 | 144 | All SZGR 2.0 genes in this pathway |
| KEGG SYSTEMIC LUPUS ERYTHEMATOSUS | 140 | 100 | All SZGR 2.0 genes in this pathway |
| KEGG ARRHYTHMOGENIC RIGHT VENTRICULAR CARDIOMYOPATHY ARVC | 76 | 59 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CELL COMMUNICATION | 120 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2 | 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME NEPHRIN INTERACTIONS | 20 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS | 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION | 208 | 138 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA REVERSIBLY DN | 29 | 21 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE UP | 134 | 93 | All SZGR 2.0 genes in this pathway |
| TERAMOTO OPN TARGETS CLUSTER 6 | 27 | 17 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| TAGHAVI NEOPLASTIC TRANSFORMATION | 12 | 10 | All SZGR 2.0 genes in this pathway |
| AMIT SERUM RESPONSE 480 MCF10A | 39 | 20 | All SZGR 2.0 genes in this pathway |
| MORI PRE BI LYMPHOCYTE UP | 80 | 54 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| MORI IMMATURE B LYMPHOCYTE DN | 90 | 55 | All SZGR 2.0 genes in this pathway |
| LIN APC TARGETS | 77 | 55 | All SZGR 2.0 genes in this pathway |
| JECHLINGER EPITHELIAL TO MESENCHYMAL TRANSITION DN | 66 | 47 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| FAELT B CLL WITH VH3 21 UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| HENDRICKS SMARCA4 TARGETS UP | 56 | 35 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS UP | 113 | 71 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS DN | 96 | 71 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| GHO ATF5 TARGETS DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| ZHONG SECRETOME OF LUNG CANCER AND FIBROBLAST | 132 | 93 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE BY GAMMA AND UV RADIATION | 88 | 65 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN | 195 | 135 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| HOFFMANN LARGE TO SMALL PRE BII LYMPHOCYTE UP | 163 | 102 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA TARGETS OF HIF1A AND FOXA2 | 37 | 27 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| DELACROIX RAR BOUND ES | 462 | 273 | All SZGR 2.0 genes in this pathway |