| REACTOME RESPONSE TO ELEVATED PLATELET CYTOSOLIC CA2
| 89 | 56 | All SZGR 2.0 genes in this pathway |
| REACTOME HEMOSTASIS
| 466 | 331 | All SZGR 2.0 genes in this pathway |
| REACTOME PLATELET ACTIVATION SIGNALING AND AGGREGATION
| 208 | 138 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP
| 423 | 283 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER CLASSES DN
| 34 | 26 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP
| 305 | 185 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP
| 204 | 140 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN
| 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP
| 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP
| 722 | 443 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS C UP
| 170 | 114 | All SZGR 2.0 genes in this pathway |
| WANG ESOPHAGUS CANCER VS NORMAL UP
| 121 | 72 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL UP
| 83 | 51 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP
| 1037 | 673 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN
| 411 | 249 | All SZGR 2.0 genes in this pathway |
| SWEET KRAS TARGETS DN
| 66 | 39 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN
| 121 | 79 | All SZGR 2.0 genes in this pathway |
| CHEN LUNG CANCER SURVIVAL
| 28 | 22 | All SZGR 2.0 genes in this pathway |
| PENG RAPAMYCIN RESPONSE DN
| 245 | 154 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP
| 555 | 346 | All SZGR 2.0 genes in this pathway |
| BHATTACHARYA EMBRYONIC STEM CELL
| 89 | 60 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND UP
| 77 | 52 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN
| 408 | 274 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS UP
| 175 | 116 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY NO BLOOD UP
| 222 | 139 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP
| 487 | 303 | All SZGR 2.0 genes in this pathway |
| JAZAERI BREAST CANCER BRCA1 VS BRCA2 UP
| 49 | 28 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN
| 434 | 302 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE
| 261 | 166 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 5
| 482 | 296 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED
| 728 | 415 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP
| 388 | 234 | All SZGR 2.0 genes in this pathway |
| KYNG DNA DAMAGE DN
| 195 | 135 | All SZGR 2.0 genes in this pathway |
| KYNG ENVIRONMENTAL STRESS RESPONSE UP
| 56 | 41 | All SZGR 2.0 genes in this pathway |
| FUJII YBX1 TARGETS DN
| 202 | 132 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP
| 973 | 570 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN
| 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP
| 648 | 398 | All SZGR 2.0 genes in this pathway |
| ITO PTTG1 TARGETS UP
| 12 | 9 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP
| 425 | 298 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA DN
| 100 | 63 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP
| 245 | 159 | All SZGR 2.0 genes in this pathway |
| BOHN PRIMARY IMMUNODEFICIENCY SYNDROM UP
| 47 | 30 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN
| 442 | 275 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN
| 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN
| 668 | 419 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER UP
| 288 | 168 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS
| 535 | 325 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP
| 605 | 377 | All SZGR 2.0 genes in this pathway |
| WIERENGA PML INTERACTOME
| 42 | 23 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN
| 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B
| 549 | 316 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE
| 1137 | 655 | All SZGR 2.0 genes in this pathway |
| MIKHAYLOVA OXIDATIVE STRESS RESPONSE VIA VHL DN
| 6 | 5 | All SZGR 2.0 genes in this pathway |
| BAKKER FOXO3 TARGETS DN
| 187 | 109 | All SZGR 2.0 genes in this pathway |
| AZARE NEOPLASTIC TRANSFORMATION BY STAT3 UP
| 121 | 70 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D
| 882 | 506 | All SZGR 2.0 genes in this pathway |
| DELACROIX RARG BOUND MEF
| 367 | 231 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS UP
| 279 | 155 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A
| 182 | 108 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE B ALL UP
| 38 | 22 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN VINCRISTINE RESISTANCE ALL UP
| 27 | 16 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP
| 489 | 314 | All SZGR 2.0 genes in this pathway |
| ZWANG EGF PERSISTENTLY UP
| 32 | 24 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY
| 1725 | 838 | All SZGR 2.0 genes in this pathway |
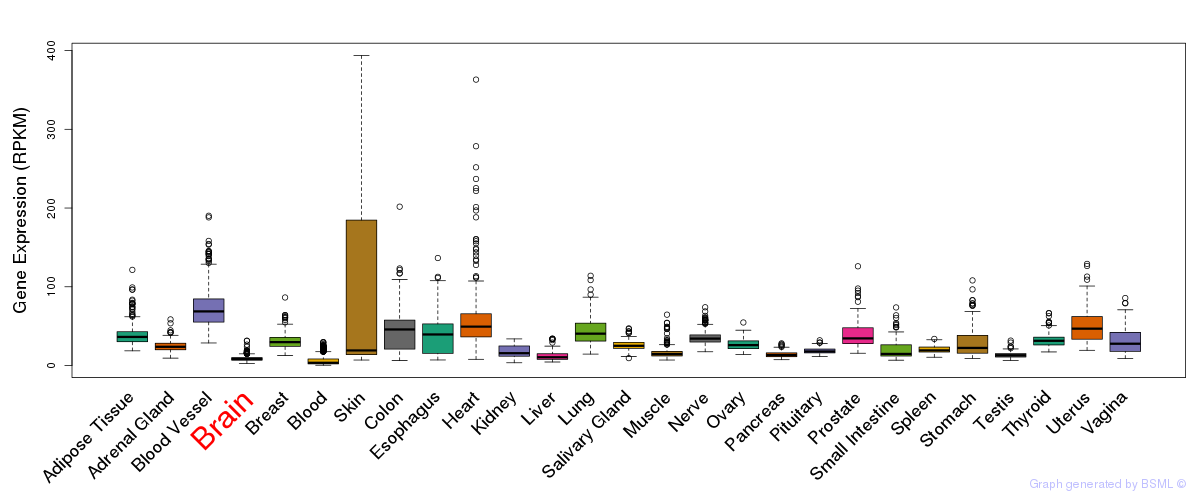
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation