Gene Page: TRIM11
Summary ?
| GeneID | 81559 |
| Symbol | TRIM11 |
| Synonyms | BIA1|RNF92 |
| Description | tripartite motif containing 11 |
| Reference | MIM:607868|HGNC:HGNC:16281|Ensembl:ENSG00000154370|HPRD:07429|Vega:OTTHUMG00000039773 |
| Gene type | protein-coding |
| Map location | 1q42.13 |
| Sherlock p-value | 0.254 |
| Fetal beta | -0.271 |
| DMG | 1 (# studies) |
| eGene | Cortex Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg17465697 | 1 | 228594029 | TRIM11 | 1.8E-6 | -0.343 | 0.008 | DMG:Wockner_2014 |
| cg07602073 | 1 | 228595680 | TRIM17;TRIM11 | 7.2E-5 | 0.475 | 0.025 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16829545 | chr2 | 151977407 | TRIM11 | 81559 | 2.923E-8 | trans | ||
| rs7584986 | chr2 | 184111432 | TRIM11 | 81559 | 0.05 | trans | ||
| rs16955618 | chr15 | 29937543 | TRIM11 | 81559 | 7.985E-12 | trans | ||
| rs1041786 | chr21 | 22617710 | TRIM11 | 81559 | 0.04 | trans |
Section II. Transcriptome annotation
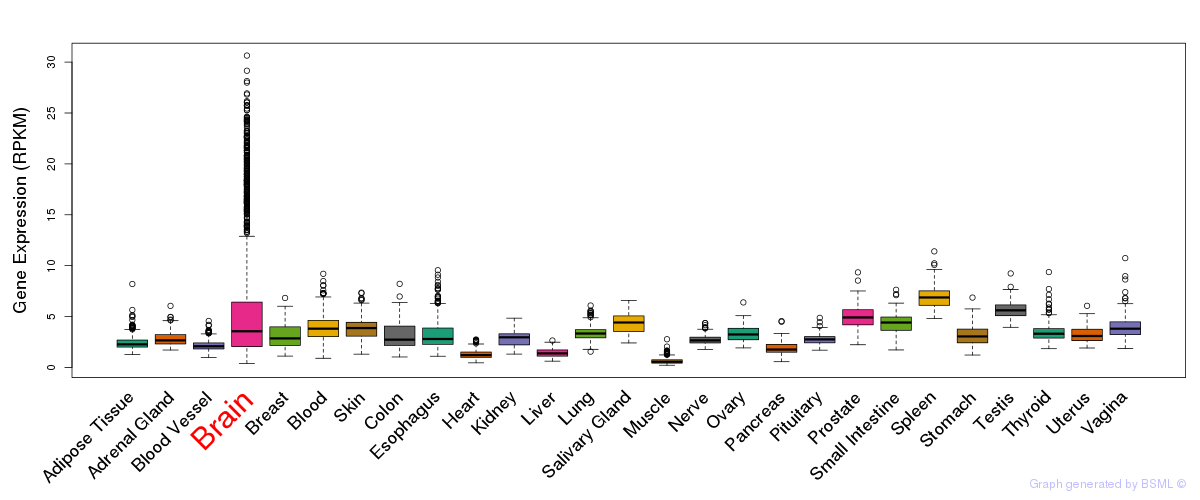
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC023055.4 | 0.38 | 0.34 |
| ACADVL | 0.34 | 0.31 |
| MOV10 | 0.34 | 0.32 |
| PLD2 | 0.33 | 0.33 |
| NPHP3 | 0.33 | 0.29 |
| USP40 | 0.33 | 0.29 |
| ATHL1 | 0.33 | 0.28 |
| PDE6B | 0.32 | 0.29 |
| MYO15B | 0.32 | 0.30 |
| DNHD1 | 0.32 | 0.31 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FAM163B | -0.27 | -0.26 |
| KCNF1 | -0.26 | -0.26 |
| LNX1 | -0.25 | -0.27 |
| HSPBP1 | -0.25 | -0.23 |
| HS3ST2 | -0.25 | -0.25 |
| CLTB | -0.25 | -0.25 |
| AC073610.6 | -0.24 | -0.26 |
| CDK5R2 | -0.24 | -0.23 |
| C1QTNF4 | -0.24 | -0.22 |
| MATK | -0.23 | -0.22 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PROCESSING UBIQUITINATION PROTEASOME DEGRADATION | 212 | 129 | All SZGR 2.0 genes in this pathway |
| CASTELLANO NRAS TARGETS UP | 68 | 41 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE UP | 86 | 57 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |