| KEGG GLYCEROPHOSPHOLIPID METABOLISM
| 77 | 35 | All SZGR 2.0 genes in this pathway |
| KEGG ETHER LIPID METABOLISM
| 33 | 15 | All SZGR 2.0 genes in this pathway |
| KEGG ARACHIDONIC ACID METABOLISM
| 58 | 36 | All SZGR 2.0 genes in this pathway |
| KEGG LINOLEIC ACID METABOLISM
| 29 | 16 | All SZGR 2.0 genes in this pathway |
| KEGG ALPHA LINOLENIC ACID METABOLISM
| 19 | 9 | All SZGR 2.0 genes in this pathway |
| KEGG MAPK SIGNALING PATHWAY
| 267 | 205 | All SZGR 2.0 genes in this pathway |
| KEGG VASCULAR SMOOTH MUSCLE CONTRACTION
| 115 | 81 | All SZGR 2.0 genes in this pathway |
| KEGG VEGF SIGNALING PATHWAY
| 76 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG FC EPSILON RI SIGNALING PATHWAY
| 79 | 58 | All SZGR 2.0 genes in this pathway |
| KEGG LONG TERM DEPRESSION
| 70 | 53 | All SZGR 2.0 genes in this pathway |
| KEGG GNRH SIGNALING PATHWAY
| 101 | 77 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PI
| 15 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PC
| 22 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHOLIPID METABOLISM
| 198 | 112 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF PA
| 27 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PG
| 16 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PE
| 21 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME ACYL CHAIN REMODELLING OF PS
| 15 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME GLYCEROPHOSPHOLIPID BIOSYNTHESIS
| 82 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS
| 478 | 302 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP
| 1382 | 904 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP
| 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP
| 450 | 256 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN
| 459 | 276 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL UP
| 316 | 190 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN
| 199 | 124 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN
| 537 | 339 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP
| 1142 | 669 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS NEUROEPITHELIUM UP
| 176 | 111 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN
| 428 | 306 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP
| 283 | 177 | All SZGR 2.0 genes in this pathway |
| MORI MATURE B LYMPHOCYTE DN
| 75 | 43 | All SZGR 2.0 genes in this pathway |
| UEDA PERIFERAL CLOCK
| 169 | 111 | All SZGR 2.0 genes in this pathway |
| MCCLUNG CREB1 TARGETS DN
| 57 | 39 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS UP
| 388 | 234 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION DN
| 60 | 38 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC
| 420 | 269 | All SZGR 2.0 genes in this pathway |
| STEIN ESRRA TARGETS
| 535 | 325 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D
| 658 | 397 | All SZGR 2.0 genes in this pathway |
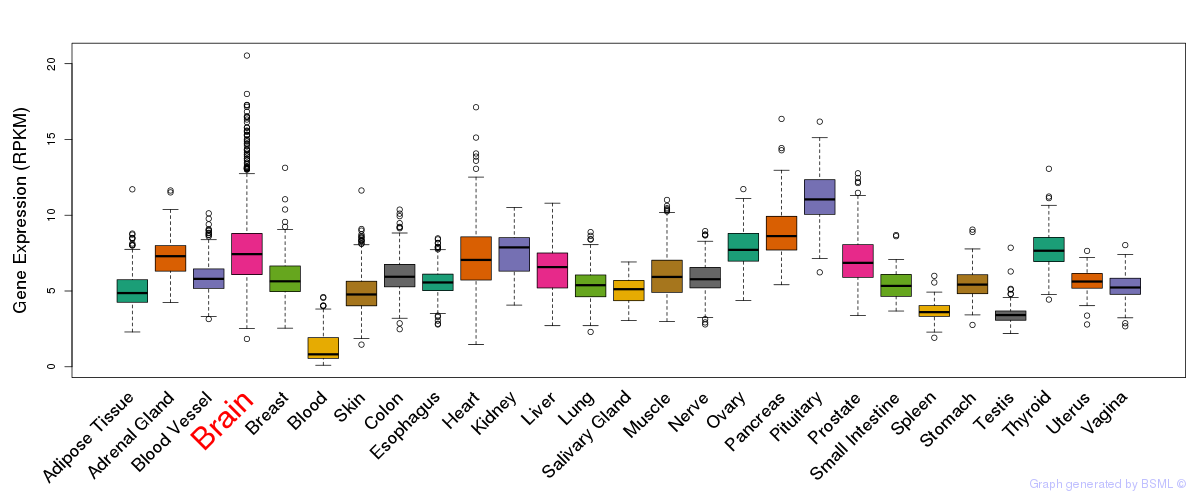
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation