Gene Page: MIA
Summary ?
| GeneID | 8190 |
| Symbol | MIA |
| Synonyms | CD-RAP |
| Description | melanoma inhibitory activity |
| Reference | MIM:601340|HGNC:HGNC:7076|Ensembl:ENSG00000261857|HPRD:03216|Vega:OTTHUMG00000182693 |
| Gene type | protein-coding |
| Map location | 19q13.2 |
| Pascal p-value | 0.311 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias,schizotypal | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
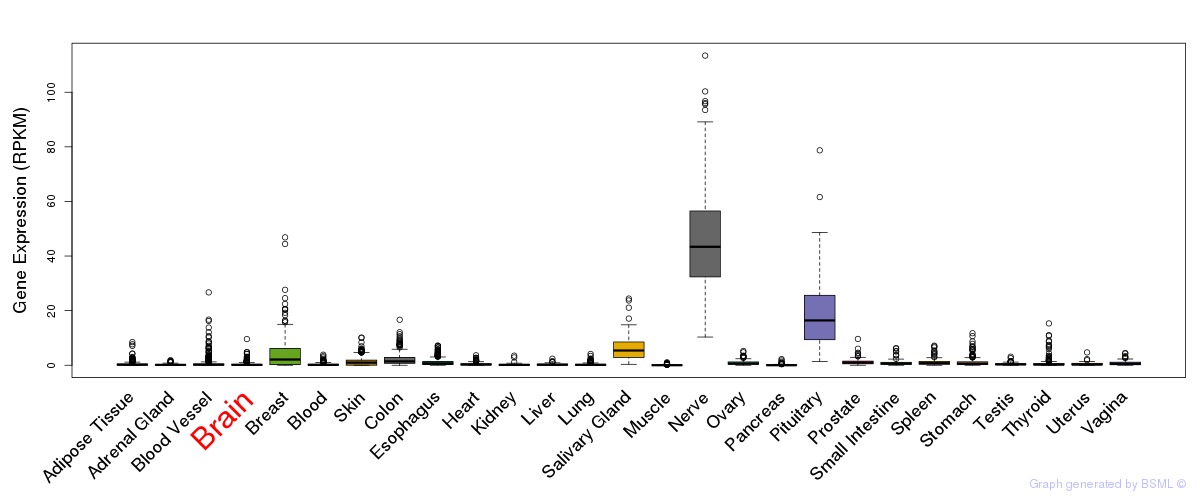
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NEURL4 | 0.95 | 0.96 |
| USP19 | 0.94 | 0.95 |
| FBXO42 | 0.93 | 0.94 |
| TSC2 | 0.93 | 0.94 |
| EDC4 | 0.93 | 0.94 |
| MBD1 | 0.92 | 0.93 |
| NLGN2 | 0.92 | 0.93 |
| ZNF142 | 0.92 | 0.94 |
| FOXK2 | 0.92 | 0.94 |
| LSM14B | 0.92 | 0.94 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.78 | -0.86 |
| MT-CO2 | -0.75 | -0.85 |
| AF347015.27 | -0.75 | -0.84 |
| AF347015.21 | -0.74 | -0.92 |
| FXYD1 | -0.74 | -0.82 |
| AF347015.33 | -0.74 | -0.81 |
| S100B | -0.73 | -0.82 |
| AC021016.1 | -0.73 | -0.83 |
| COPZ2 | -0.73 | -0.79 |
| AF347015.8 | -0.72 | -0.83 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER ESR1 DN | 48 | 28 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| AUNG GASTRIC CANCER | 54 | 20 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR UP | 487 | 286 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| ABE INNER EAR | 48 | 26 | All SZGR 2.0 genes in this pathway |
| SATO SILENCED BY METHYLATION IN PANCREATIC CANCER 1 | 419 | 273 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| HARRIS BRAIN CANCER PROGENITORS | 44 | 23 | All SZGR 2.0 genes in this pathway |
| IWANAGA CARCINOGENESIS BY KRAS PTEN UP | 181 | 112 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN PLEURA DN | 27 | 15 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A UP | 104 | 57 | All SZGR 2.0 genes in this pathway |
| HUPER BREAST BASAL VS LUMINAL DN | 59 | 44 | All SZGR 2.0 genes in this pathway |
| CROONQUIST NRAS VS STROMAL STIMULATION UP | 41 | 26 | All SZGR 2.0 genes in this pathway |
| POOLA INVASIVE BREAST CANCER DN | 134 | 83 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL UP | 489 | 314 | All SZGR 2.0 genes in this pathway |