Gene Page: CANX
Summary ?
| GeneID | 821 |
| Symbol | CANX |
| Synonyms | CNX|IP90|P90 |
| Description | calnexin |
| Reference | MIM:114217|HGNC:HGNC:1473|Ensembl:ENSG00000127022|HPRD:00252|Vega:OTTHUMG00000130910 |
| Gene type | protein-coding |
| Map location | 5q35 |
| Pascal p-value | 0.105 |
| Sherlock p-value | 0.197 |
| Fetal beta | -0.129 |
| eGene | Cortex Meta |
| Support | RNA AND PROTEIN SYNTHESIS CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| GSMA_IIA | Genome scan meta-analysis (All samples) | Psr: 0.0276 |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| CANX | chr5 | 179136974 | A | G | NM_001024649 NM_001746 | p.210K>R p.210K>R | missense missense | Schizophrenia | DNM:Xu_2012 |
Section II. Transcriptome annotation
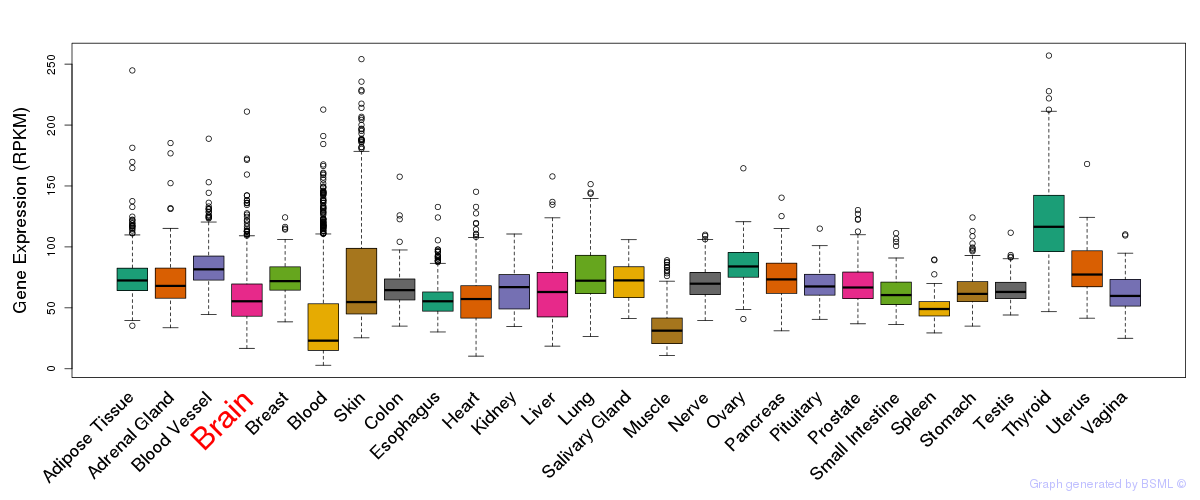
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GJB1 | 0.95 | 0.90 |
| KLK6 | 0.94 | 0.90 |
| SLC5A11 | 0.93 | 0.93 |
| MOG | 0.93 | 0.91 |
| TF | 0.92 | 0.92 |
| SLC31A2 | 0.92 | 0.82 |
| PLA2G16 | 0.92 | 0.91 |
| DBNDD2 | 0.92 | 0.90 |
| PPAP2C | 0.91 | 0.85 |
| SGK2 | 0.91 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NKIRAS2 | -0.55 | -0.71 |
| CRMP1 | -0.53 | -0.69 |
| TUBB2B | -0.53 | -0.78 |
| HN1 | -0.52 | -0.71 |
| HMGB3 | -0.52 | -0.72 |
| KIAA1949 | -0.52 | -0.71 |
| TUBB | -0.52 | -0.72 |
| YBX1 | -0.52 | -0.78 |
| PCDHB18 | -0.51 | -0.70 |
| CD24 | -0.51 | -0.71 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005529 | sugar binding | IEA | - | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005509 | calcium ion binding | TAS | 8136357 | |
| GO:0051082 | unfolded protein binding | IEA | - | |
| GO:0051082 | unfolded protein binding | NAS | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001525 | angiogenesis | IDA | 15467828 | |
| GO:0006457 | protein folding | IEA | - | |
| GO:0009306 | protein secretion | TAS | 8055875 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005789 | endoplasmic reticulum membrane | IEA | - | |
| GO:0005737 | cytoplasm | IDA | 15467828 | |
| GO:0005783 | endoplasmic reticulum | IDA | 14966132 |15467828 | |
| GO:0005783 | endoplasmic reticulum | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0042470 | melanosome | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABCC2 | ABC30 | CMOAT | DJS | KIAA1010 | MRP2 | cMRP | ATP-binding cassette, sub-family C (CFTR/MRP), member 2 | - | HPRD | 12222674 |
| APOB | FLDB | apolipoprotein B (including Ag(x) antigen) | Affinity Capture-Western | BioGRID | 14498830 |
| BCAP31 | 6C6-AG | BAP31 | CDM | DXS1357E | B-cell receptor-associated protein 31 | - | HPRD | 11851433 |
| CALR | CRT | FLJ26680 | RO | SSA | cC1qR | calreticulin | Affinity Capture-Western | BioGRID | 10436013 |
| CD1D | CD1A | MGC34622 | R3 | CD1d molecule | - | HPRD,BioGRID | 12239218 |
| CD3D | CD3-DELTA | T3D | CD3d molecule, delta (CD3-TCR complex) | - | HPRD,BioGRID | 8278814 |8621641 |
| CD82 | 4F9 | C33 | GR15 | IA4 | KAI1 | R2 | SAR2 | ST6 | TSPAN27 | CD82 molecule | - | HPRD | 11350933 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | CFTR interacts with CANX. This interaction was modeled on a demonstrated interaction between human CFTR and hamster CANX. | BIND | 15105504 |
| CFTR | ABC35 | ABCC7 | CF | CFTR/MRP | MRP7 | TNR-CFTR | dJ760C5.1 | cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) | CFTR interacts with CANX. | BIND | 7513695 |
| EBI3 | IL27B | Epstein-Barr virus induced 3 | - | HPRD,BioGRID | 8551575 |
| EDEM1 | EDEM | FLJ51559 | FLJ51560 | KIAA0212 | ER degradation enhancer, mannosidase alpha-like 1 | - | HPRD,BioGRID | 12610305 |
| F8 | AHF | DXS1253E | F8B | F8C | FVIII | HEMA | coagulation factor VIII, procoagulant component | - | HPRD,BioGRID | 9525969 |
| GRIA1 | GLUH1 | GLUR1 | GLURA | HBGR1 | MGC133252 | glutamate receptor, ionotropic, AMPA 1 | - | HPRD | 10461883 |
| GRIN1 | NMDA1 | NMDAR1 | NR1 | glutamate receptor, ionotropic, N-methyl D-aspartate 1 | - | HPRD | 11506858 |14732708 |
| HTR3A | 5-HT-3 | 5-HT3A | 5-HT3R | 5HT3R | HTR3 | 5-hydroxytryptamine (serotonin) receptor 3A | - | HPRD | 12359150 |
| ITGA6 | CD49f | DKFZp686J01244 | FLJ18737 | ITGA6B | VLA-6 | integrin, alpha 6 | - | HPRD | 8163531 |
| ITGB1 | CD29 | FNRB | GPIIA | MDF2 | MSK12 | VLA-BETA | VLAB | integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12) | - | HPRD,BioGRID | 8163531 |
| LCT | LAC | LPH | LPH1 | lactase | - | HPRD,BioGRID | 11751874 |
| LPA | AK38 | APOA | LP | lipoprotein, Lp(a) | - | HPRD,BioGRID | 9717723 |12562843 |
| LRP1 | A2MR | APOER | APR | CD91 | FLJ16451 | IGFBP3R | LRP | MGC88725 | TGFBR5 | low density lipoprotein-related protein 1 (alpha-2-macroglobulin receptor) | - | HPRD,BioGRID | 14980518 |
| MBTPS1 | KIAA0091 | MGC138711 | MGC138712 | PCSK8 | S1P | SKI-1 | membrane-bound transcription factor peptidase, site 1 | - | HPRD | 11756446 |
| PDIA3 | ER60 | ERp57 | ERp60 | ERp61 | GRP57 | GRP58 | HsT17083 | P58 | PI-PLC | protein disulfide isomerase family A, member 3 | ERp57 interacts with CNX. This interaction was modeled on a demonstrated interaction between dog CNX and ERp57 from an unspecified source. | BIND | 12052826 |
| PDIA3 | ER60 | ERp57 | ERp60 | ERp61 | GRP57 | GRP58 | HsT17083 | P58 | PI-PLC | protein disulfide isomerase family A, member 3 | - | HPRD | 10436013 |
| PIGK | GPI8 | MGC22559 | phosphatidylinositol glycan anchor biosynthesis, class K | Affinity Capture-MS | BioGRID | 12582175 |
| PRKCSH | AGE-R2 | G19P1 | PCLD | PLD1 | protein kinase C substrate 80K-H | - | HPRD | 8910335 |
| SERP1 | MGC117327 | MGC133321 | MGC133322 | RAMP4 | stress-associated endoplasmic reticulum protein 1 | - | HPRD | 10601334 |
| SLC2A1 | DYT17 | DYT18 | GLUT | GLUT1 | MGC141895 | MGC141896 | PED | solute carrier family 2 (facilitated glucose transporter), member 1 | - | HPRD,BioGRID | 8662691 |
| SLC4A1 | AE1 | BND3 | CD233 | DI | EMPB3 | EPB3 | FR | MGC116750 | MGC116753 | MGC126619 | MGC126623 | RTA1A | SW | WD | WD1 | WR | solute carrier family 4, anion exchanger, member 1 (erythrocyte membrane protein band 3, Diego blood group) | - | HPRD,BioGRID | 10364201 |
| SLC6A4 | 5-HTT | 5HTT | HTT | OCD1 | SERT | hSERT | solute carrier family 6 (neurotransmitter transporter, serotonin), member 4 | Reconstituted Complex | BioGRID | 10364189 |
| TF | DKFZp781D0156 | PRO1557 | PRO2086 | transferrin | - | HPRD,BioGRID | 9312001 |
| TG | AITD3 | TGN | thyroglobulin | - | HPRD | 10636893 |
| TRA@ | FLJ22602 | MGC117436 | MGC22624 | MGC23964 | MGC71411 | TCRA | TCRD | TRA | T cell receptor alpha locus | - | HPRD | 11566319 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | Thyroid-Stimulating Hormone Receptor interacts with the molecular chaperone calnexin. This interaction was modeled on a demonstrated interaction between human TSHR and dog calnexin. | BIND | 12383251 |
| TSHR | CHNG1 | LGR3 | MGC75129 | hTSHR-I | thyroid stimulating hormone receptor | - | HPRD,BioGRID | 12383251 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ANTIGEN PROCESSING AND PRESENTATION | 89 | 65 | All SZGR 2.0 genes in this pathway |
| REACTOME MHC CLASS II ANTIGEN PRESENTATION | 91 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME ASPARAGINE N LINKED GLYCOSYLATION | 81 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME CALNEXIN CALRETICULIN CYCLE | 11 | 7 | All SZGR 2.0 genes in this pathway |
| REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | 13 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLUENZA LIFE CYCLE | 203 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | 21 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME CLASS I MHC MEDIATED ANTIGEN PROCESSING PRESENTATION | 251 | 156 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| PRAMOONJAGO SOX4 TARGETS DN | 51 | 35 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS DN | 310 | 188 | All SZGR 2.0 genes in this pathway |
| RHEIN ALL GLUCOCORTICOID THERAPY DN | 362 | 238 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR DN | 214 | 133 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES DN | 16 | 10 | All SZGR 2.0 genes in this pathway |
| LI AMPLIFIED IN LUNG CANCER | 178 | 108 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| TOMLINS PROSTATE CANCER UP | 40 | 27 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT AND CANCER BOX4 DN | 32 | 22 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP | 78 | 51 | All SZGR 2.0 genes in this pathway |
| GOLDRATH HOMEOSTATIC PROLIFERATION | 171 | 102 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR DN | 41 | 30 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT REJECTED VS OK UP | 63 | 48 | All SZGR 2.0 genes in this pathway |
| NGUYEN NOTCH1 TARGETS UP | 29 | 23 | All SZGR 2.0 genes in this pathway |
| MUNSHI MULTIPLE MYELOMA UP | 81 | 52 | All SZGR 2.0 genes in this pathway |
| RHODES CANCER META SIGNATURE | 64 | 47 | All SZGR 2.0 genes in this pathway |
| JIANG AGING HYPOTHALAMUS DN | 40 | 31 | All SZGR 2.0 genes in this pathway |
| JIANG VHL TARGETS | 138 | 91 | All SZGR 2.0 genes in this pathway |
| PAL PRMT5 TARGETS DN | 29 | 16 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA NORMAL | 311 | 205 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| PELLICCIOTTA HDAC IN ANTIGEN PRESENTATION DN | 49 | 38 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| GRADE COLON VS RECTAL CANCER UP | 38 | 21 | All SZGR 2.0 genes in this pathway |
| CUI GLUCOSE DEPRIVATION | 60 | 44 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| WANG TUMOR INVASIVENESS DN | 210 | 128 | All SZGR 2.0 genes in this pathway |
| BOHN PRIMARY IMMUNODEFICIENCY SYNDROM UP | 47 | 30 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 MULTIPLE MYELOMA PROGRAM | 36 | 25 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN MYELOMA VS MATURE B LYMPHOCYTE | 101 | 76 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN PLASMA CELL VS MATURE B LYMPHOCYTE | 67 | 51 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED B LYMPHOCYTE | 81 | 66 | All SZGR 2.0 genes in this pathway |
| SHAFFER IRF4 TARGETS IN ACTIVATED DENDRITIC CELL | 65 | 49 | All SZGR 2.0 genes in this pathway |
| HSIAO HOUSEKEEPING GENES | 389 | 245 | All SZGR 2.0 genes in this pathway |
| ROME INSULIN TARGETS IN MUSCLE UP | 442 | 263 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| GUTIERREZ MULTIPLE MYELOMA UP | 35 | 24 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 10 | 69 | 38 | All SZGR 2.0 genes in this pathway |
| KYNG RESPONSE TO H2O2 VIA ERCC6 UP | 40 | 30 | All SZGR 2.0 genes in this pathway |
| JIANG HYPOXIA VIA VHL | 34 | 24 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-128 | 858 | 864 | m8 | hsa-miR-128a | UCACAGUGAACCGGUCUCUUUU |
| hsa-miR-128b | UCACAGUGAACCGGUCUCUUUC | ||||
| miR-130/301 | 1543 | 1549 | m8 | hsa-miR-130abrain | CAGUGCAAUGUUAAAAGGGCAU |
| hsa-miR-301 | CAGUGCAAUAGUAUUGUCAAAGC | ||||
| hsa-miR-130bbrain | CAGUGCAAUGAUGAAAGGGCAU | ||||
| hsa-miR-454-3p | UAGUGCAAUAUUGCUUAUAGGGUUU | ||||
| miR-139 | 204 | 210 | 1A | hsa-miR-139brain | UCUACAGUGCACGUGUCU |
| miR-148/152 | 1544 | 1551 | 1A,m8 | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-153 | 918 | 924 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-205 | 1616 | 1622 | m8 | hsa-miR-205 | UCCUUCAUUCCACCGGAGUCUG |
| miR-29 | 1403 | 1409 | 1A | hsa-miR-29aSZ | UAGCACCAUCUGAAAUCGGUU |
| hsa-miR-29bSZ | UAGCACCAUUUGAAAUCAGUGUU | ||||
| hsa-miR-29cSZ | UAGCACCAUUUGAAAUCGGU | ||||
| miR-30-3p | 1461 | 1467 | m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC | ||||
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-374 | 965 | 971 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-494 | 1009 | 1015 | m8 | hsa-miR-494brain | UGAAACAUACACGGGAAACCUCUU |
| miR-496 | 1280 | 1286 | m8 | hsa-miR-496 | AUUACAUGGCCAAUCUC |
| miR-505 | 864 | 870 | m8 | hsa-miR-505 | GUCAACACUUGCUGGUUUCCUC |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.