Gene Page: DYSF
Summary ?
| GeneID | 8291 |
| Symbol | DYSF |
| Synonyms | FER1L1|LGMD2B|MMD1 |
| Description | dysferlin |
| Reference | MIM:603009|HGNC:HGNC:3097|Ensembl:ENSG00000135636|HPRD:04307|Vega:OTTHUMG00000129757 |
| Gene type | protein-coding |
| Map location | 2p13.3 |
| Pascal p-value | 0.092 |
| Sherlock p-value | 0.25 |
| Fetal beta | -0.849 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg03170698 | 2 | 71694016 | DYSF | 1.224E-4 | -0.464 | 0.03 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6829546 | chr4 | 12587664 | DYSF | 8291 | 0.19 | trans | ||
| rs1465923 | chr4 | 77136210 | DYSF | 8291 | 0.19 | trans |
Section II. Transcriptome annotation
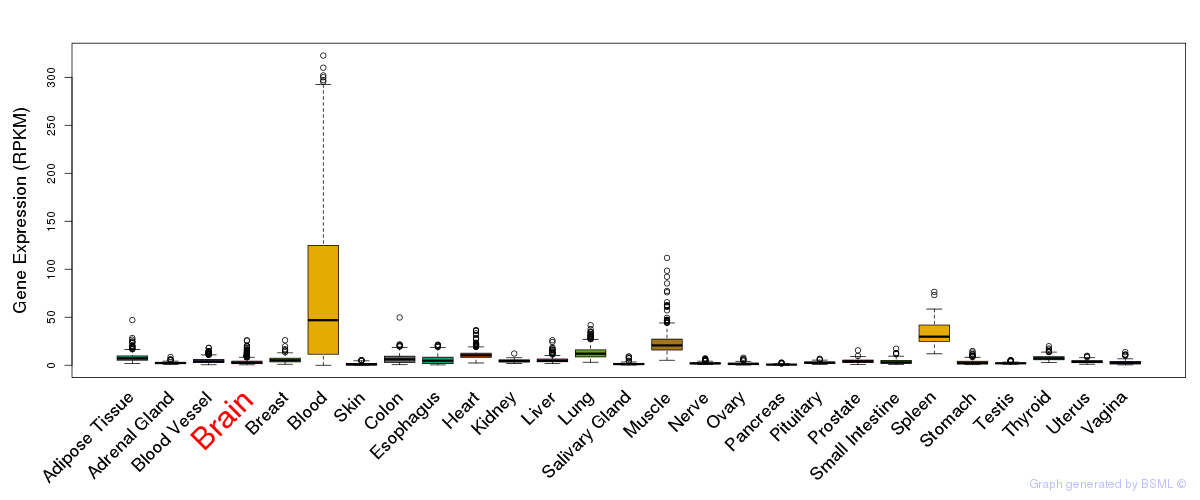
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HIST1H2BD | 0.92 | 0.77 |
| KDM3A | 0.85 | 0.62 |
| TMCO7 | 0.81 | 0.62 |
| HMGA1 | 0.80 | 0.64 |
| PCBP2 | 0.79 | 0.66 |
| ZNF143 | 0.79 | 0.64 |
| BBS9 | 0.79 | 0.65 |
| SOX4 | 0.79 | 0.63 |
| USP3 | 0.79 | 0.68 |
| H3F3B | 0.78 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C5orf53 | -0.53 | -0.63 |
| FXYD1 | -0.47 | -0.61 |
| IFI27 | -0.46 | -0.56 |
| MT-CO2 | -0.46 | -0.58 |
| MUSTN1 | -0.45 | -0.52 |
| AC018755.7 | -0.45 | -0.50 |
| AF347015.31 | -0.45 | -0.55 |
| HIGD1B | -0.44 | -0.58 |
| AF347015.27 | -0.44 | -0.56 |
| S100B | -0.44 | -0.53 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 UP | 146 | 86 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS F UP | 185 | 119 | All SZGR 2.0 genes in this pathway |
| HATADA METHYLATED IN LUNG CANCER UP | 390 | 236 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 DN | 74 | 40 | All SZGR 2.0 genes in this pathway |
| DELASERNA MYOD TARGETS UP | 89 | 51 | All SZGR 2.0 genes in this pathway |
| YAMASHITA METHYLATED IN PROSTATE CANCER | 57 | 29 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K9ME3 DN | 120 | 71 | All SZGR 2.0 genes in this pathway |
| VANTVEER BREAST CANCER ESR1 DN | 240 | 153 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |