Gene Page: TRRAP
Summary ?
| GeneID | 8295 |
| Symbol | TRRAP |
| Synonyms | PAF350/400|PAF400|STAF40|TR-AP|Tra1 |
| Description | transformation/transcription domain-associated protein |
| Reference | MIM:603015|HGNC:HGNC:12347|Ensembl:ENSG00000196367|HPRD:04310|Vega:OTTHUMG00000150403 |
| Gene type | protein-coding |
| Map location | 7q21.2-q22.1 |
| Pascal p-value | 0.451 |
| Sherlock p-value | 0.496 |
| TADA p-value | 0.036 |
| Support | CompositeSet Darnell FMRP targets Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Xu_2012 | Whole Exome Sequencing analysis | De novo mutations of 4 genes were identified by exome sequencing of 795 samples in this study | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| TRRAP | chr7 | 98498329 | A | T | NM_001244580 NM_003496 | p.295I>F p.295I>F | missense missense | Schizophrenia | DNM:Xu_2012 |
Section II. Transcriptome annotation
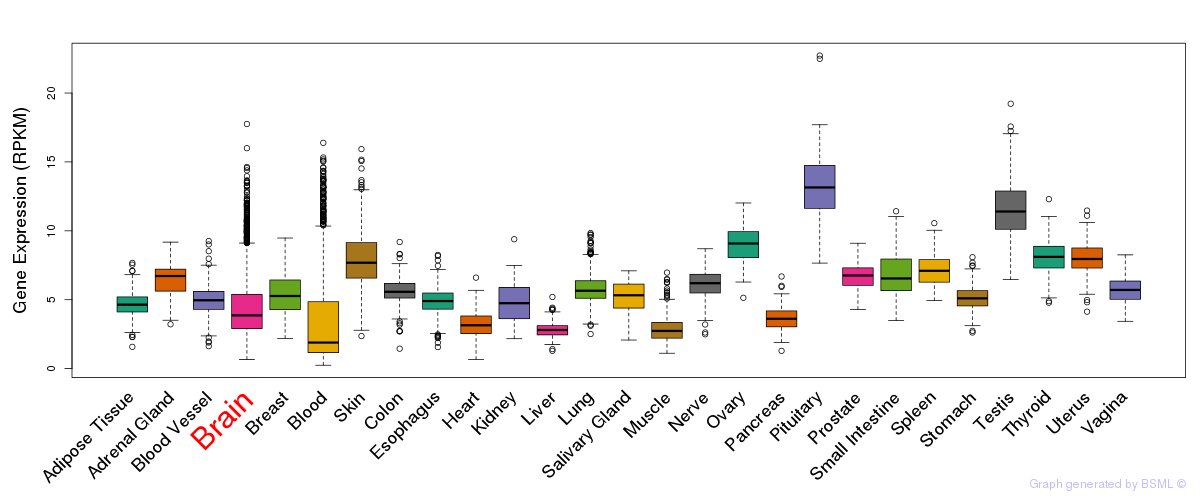
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HIST1H3H | 0.65 | 0.25 |
| HIST1H2AM | 0.61 | 0.20 |
| NEK2 | 0.61 | 0.40 |
| CENPN | 0.61 | 0.47 |
| CDC25B | 0.59 | 0.52 |
| AURKA | 0.59 | 0.31 |
| HIST2H2AC | 0.59 | 0.15 |
| EXO1 | 0.59 | 0.23 |
| DSN1 | 0.58 | 0.45 |
| HIST1H2BJ | 0.58 | 0.26 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EMID1 | -0.22 | -0.26 |
| CCBE1 | -0.21 | -0.09 |
| SATB2 | -0.20 | -0.04 |
| SYCP3 | -0.19 | -0.18 |
| RPRM | -0.19 | -0.08 |
| CLDN10 | -0.19 | -0.10 |
| TLCD1 | -0.18 | -0.10 |
| ADAMTSL3 | -0.18 | -0.07 |
| AF347015.21 | -0.18 | -0.11 |
| AC087071.1 | -0.18 | -0.23 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ACTL6A | ACTL6 | Arp4 | BAF53A | INO80K | MGC5382 | actin-like 6A | - | HPRD,BioGRID | 11839798 |
| C20orf20 | Eaf7 | FLJ10914 | MRG15BP | MRGBP | URCC4 | chromosome 20 open reading frame 20 | - | HPRD | 12963728 |
| E2F1 | E2F-1 | RBAP1 | RBBP3 | RBP3 | E2F transcription factor 1 | - | HPRD,BioGRID | 9708738 |
| EP400 | CAGH32 | DKFZP434I225 | FLJ42018 | FLJ45115 | P400 | TNRC12 | E1A binding protein p400 | - | HPRD,BioGRID | 11509179 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | ESR1 (ER-alpha) interacts with TRRAP. This interaction was modeled on a demonstrated interaction between ESR1 from an unspecified species and TRRAP from an unspecified species. | BIND | 12738788 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | Affinity Capture-MS | BioGRID | 12837248 |
| ING1 | p24ING1c | p33 | p33ING1 | p33ING1b | p47 | p47ING1a | inhibitor of growth family, member 1 | Affinity Capture-Western | BioGRID | 12015309 |
| KAT2A | GCN5 | GCN5L2 | MGC102791 | PCAF-b | hGCN5 | K(lysine) acetyltransferase 2A | Affinity Capture-Western | BioGRID | 10611234 |
| KAT2B | CAF | P | P/CAF | PCAF | K(lysine) acetyltransferase 2B | Affinity Capture-Western | BioGRID | 11509179 |12660246 |
| MAX | MGC10775 | MGC11225 | MGC18164 | MGC34679 | MGC36767 | bHLHd4 | orf1 | MYC associated factor X | - | HPRD,BioGRID | 10611234 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | - | HPRD,BioGRID | 9708738 |
| MYC | bHLHe39 | c-Myc | v-myc myelocytomatosis viral oncogene homolog (avian) | c-Myc interacts with TRRAP. | BIND | 12660246 |
| RUVBL1 | ECP54 | INO80H | NMP238 | PONTIN | Pontin52 | RVB1 | TIH1 | TIP49 | TIP49A | RuvB-like 1 (E. coli) | Affinity Capture-Western | BioGRID | 11839798 |
| TAL1 | SCL | TCL5 | bHLHa17 | tal-1 | T-cell acute lymphocytic leukemia 1 | Affinity Capture-MS | BioGRID | 16407974 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Affinity Capture-MS | BioGRID | 12837248 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA PITX2 PATHWAY | 15 | 15 | All SZGR 2.0 genes in this pathway |
| PID E2F PATHWAY | 74 | 48 | All SZGR 2.0 genes in this pathway |
| PID MYC ACTIV PATHWAY | 79 | 62 | All SZGR 2.0 genes in this pathway |
| PID MYC PATHWAY | 25 | 22 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID BETA CATENIN NUC PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| BILBAN B CLL LPL UP | 63 | 39 | All SZGR 2.0 genes in this pathway |
| NAGY TFTC COMPONENTS HUMAN | 19 | 11 | All SZGR 2.0 genes in this pathway |
| NAGY STAGA COMPONENTS HUMAN | 15 | 8 | All SZGR 2.0 genes in this pathway |
| NAGY PCAF COMPONENTS HUMAN | 9 | 7 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7Q21 Q22 AMPLICON | 76 | 33 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| LI CYTIDINE ANALOGS CYCTOTOXICITY | 15 | 8 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR DN | 354 | 216 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN DN | 249 | 165 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER UP | 298 | 174 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |