Gene Page: CAPZB
Summary ?
| GeneID | 832 |
| Symbol | CAPZB |
| Synonyms | CAPB|CAPPB|CAPZ |
| Description | capping actin protein of muscle Z-line beta subunit |
| Reference | MIM:601572|HGNC:HGNC:1491|Ensembl:ENSG00000077549|HPRD:11872|Vega:OTTHUMG00000002556 |
| Gene type | protein-coding |
| Map location | 1p36.1 |
| Pascal p-value | 0.088 |
| Sherlock p-value | 0.121 |
| Fetal beta | -2.141 |
| Support | STRUCTURAL PLASTICITY G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.humanNRC CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
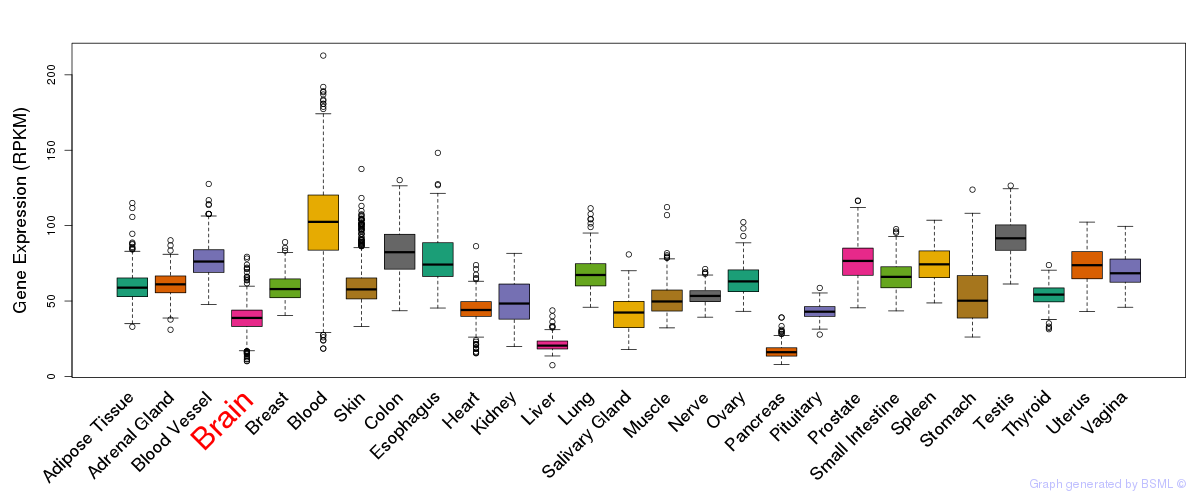
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GSPT2 | 0.89 | 0.91 |
| TTC13 | 0.89 | 0.91 |
| RARS2 | 0.89 | 0.89 |
| EIF4G2 | 0.89 | 0.92 |
| COPB1 | 0.88 | 0.91 |
| HISPPD1 | 0.88 | 0.90 |
| LRRC40 | 0.88 | 0.90 |
| SCAND3 | 0.88 | 0.90 |
| BIRC2 | 0.88 | 0.92 |
| SMARCA1 | 0.88 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HLA-F | -0.72 | -0.81 |
| AF347015.33 | -0.69 | -0.85 |
| MT-CO2 | -0.69 | -0.86 |
| AF347015.27 | -0.69 | -0.83 |
| AF347015.31 | -0.68 | -0.83 |
| MT-CYB | -0.67 | -0.84 |
| FXYD1 | -0.67 | -0.83 |
| AF347015.8 | -0.66 | -0.85 |
| TINAGL1 | -0.66 | -0.76 |
| TSC22D4 | -0.66 | -0.78 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GAZDA DIAMOND BLACKFAN ANEMIA ERYTHROID DN | 493 | 298 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| DAIRKEE TERT TARGETS UP | 380 | 213 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| NING CHRONIC OBSTRUCTIVE PULMONARY DISEASE DN | 121 | 79 | All SZGR 2.0 genes in this pathway |
| FLECHNER PBL KIDNEY TRANSPLANT OK VS DONOR UP | 151 | 100 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| KEEN RESPONSE TO ROSIGLITAZONE DN | 106 | 68 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS DN | 543 | 317 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS UP | 602 | 364 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS UP | 601 | 369 | All SZGR 2.0 genes in this pathway |
| SARRIO EPITHELIAL MESENCHYMAL TRANSITION DN | 154 | 101 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| NUTT GBM VS AO GLIOMA UP | 46 | 34 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |