Gene Page: CASP1
Summary ?
| GeneID | 834 |
| Symbol | CASP1 |
| Synonyms | ICE|IL1BC|P45 |
| Description | caspase 1 |
| Reference | MIM:147678|HGNC:HGNC:1499|Ensembl:ENSG00000137752|HPRD:00977|Vega:OTTHUMG00000048072 |
| Gene type | protein-coding |
| Map location | 11q23 |
| Pascal p-value | 0.046 |
| DEG p-value | DEG:Sanders_2014:DS1_p=0.143:DS1_beta=0.049100:DS2_p=4.63e-02:DS2_beta=-0.097:DS2_FDR=1.89e-01 |
| Fetal beta | -0.289 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DEG:Sanders_2013 | Microarray | Whole-genome gene expression profiles using microarrays on lymphoblastoid cell lines (LCLs) from 413 cases and 446 controls. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 2.5585 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
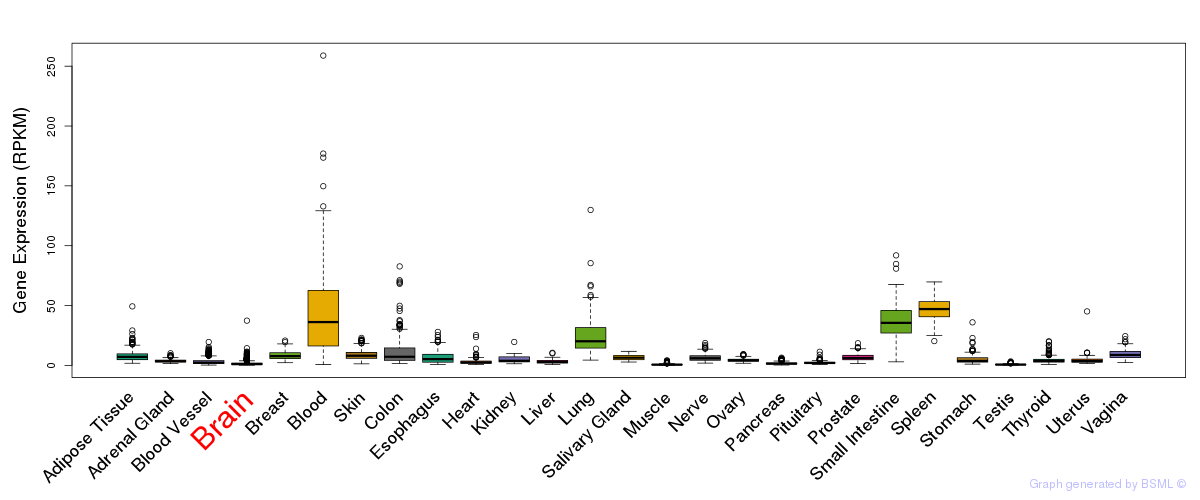
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 12888622 |15107016 | |
| GO:0004197 | cysteine-type endopeptidase activity | IEA | - | |
| GO:0004197 | cysteine-type endopeptidase activity | TAS | 10353249 | |
| GO:0008656 | caspase activator activity | TAS | 10799503 | |
| GO:0008233 | peptidase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0001666 | response to hypoxia | IEA | - | |
| GO:0006508 | proteolysis | IDA | 12888622 | |
| GO:0006508 | proteolysis | IEA | - | |
| GO:0008219 | cell death | IEA | - | |
| GO:0006917 | induction of apoptosis | IEA | - | |
| GO:0007165 | signal transduction | TAS | 1373520 | |
| GO:0042981 | regulation of apoptosis | IEA | - | |
| GO:0016485 | protein processing | IEA | - | |
| GO:0043123 | positive regulation of I-kappaB kinase/NF-kappaB cascade | IEP | 12761501 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | IEA | - | |
| GO:0005622 | intracellular | IEA | - | |
| GO:0005634 | nucleus | IDA | 18029348 | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Androgen Receptor interacts with caspase 1. This interaction was modeled on a demonstrated interaction between human androgen receptor and caspase 1 from an unspecified species. | BIND | 9535906 |
| ATN1 | B37 | D12S755E | DRPLA | NOD | atrophin 1 | Atrophin-1 interacts with caspase 1. This interaction was modeled on a demonstrated interaction between human atrophin-1 and caspase 1 from an unspecified species. | BIND | 9535906 |
| ATXN3 | AT3 | ATX3 | JOS | MJD | MJD1 | SCA3 | ataxin 3 | Ataxin-3 interacts with caspase 1. This interaction was modeled on a demonstrated interaction between human ataxin-3 and caspase 1 from an unspecified species. | BIND | 9535906 |
| ATXN3 | AT3 | ATX3 | JOS | MJD | MJD1 | SCA3 | ataxin 3 | - | HPRD | 9535906 |
| BCAP31 | 6C6-AG | BAP31 | CDM | DXS1357E | B-cell receptor-associated protein 31 | - | HPRD,BioGRID | 9334338 |
| CARD16 | COP | COP1 | PSEUDO-ICE | caspase recruitment domain family, member 16 | - | HPRD | 11432859 |11536016 |
| CARD17 | INCA | caspase recruitment domain family, member 17 | - | HPRD | 15383541 |
| CARD8 | CARDINAL | DACAR | DKFZp779L0366 | Dakar | FLJ18119 | FLJ18121 | KIAA0955 | MGC57162 | NDPP | NDPP1 | TUCAN | caspase recruitment domain family, member 8 | - | HPRD | 11821383 |
| CARD8 | CARDINAL | DACAR | DKFZp779L0366 | Dakar | FLJ18119 | FLJ18121 | KIAA0955 | MGC57162 | NDPP | NDPP1 | TUCAN | caspase recruitment domain family, member 8 | CARD-8 interacts with Casp-1 | BIND | 11821383 |
| CASP1 | ICE | IL1BC | P45 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | - | HPRD | 9190289 |
| CASP1 | ICE | IL1BC | P45 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | Affinity Capture-Western | BioGRID | 11432859 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 8962078 |
| CASP14 | MGC119078 | MGC119079 | caspase 14, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 9792675 |
| CASP4 | ICE(rel)II | ICEREL-II | ICH-2 | Mih1/TX | TX | caspase 4, apoptosis-related cysteine peptidase | Reconstituted Complex | BioGRID | 10477277 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 8962078 |
| COP | CLTO | clathrin-ordered protein | Affinity Capture-Western | BioGRID | 11432859 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD,BioGRID | 11226410 |
| HTT | HD | IT15 | huntingtin | Huntingtin interacts with caspase 1. This interaction was modeled on a demonstrated interaction between human huntingtin and caspase 1 from an unspecified species. | BIND | 9535906 |
| HTT | HD | IT15 | huntingtin | - | HPRD | 9535906 |
| IL18 | IGIF | IL-18 | IL-1g | IL1F4 | MGC12320 | interleukin 18 (interferon-gamma-inducing factor) | Biochemical Activity | BioGRID | 12096920 |
| IL1F7 | FIL1 | FIL1(ZETA) | FIL1Z | IL-1F7 | IL-1H4 | IL-1RP1 | IL1H4 | IL1RP1 | interleukin 1 family, member 7 (zeta) | Biochemical Activity | BioGRID | 12096920 |
| NA | NAC | neurocanthocytosis | Affinity Capture-Western | BioGRID | 15107016 |
| NFE2L2 | NRF2 | nuclear factor (erythroid-derived 2)-like 2 | - | HPRD,BioGRID | 10510468 |
| NLRC4 | CARD12 | CLAN | CLAN1 | CLANA | CLANB | CLANC | CLAND | CLR2.1 | IPAF | NLR family, CARD domain containing 4 | - | HPRD,BioGRID | 11472070 |
| NLRC4 | CARD12 | CLAN | CLAN1 | CLANA | CLANB | CLANC | CLAND | CLR2.1 | IPAF | NLR family, CARD domain containing 4 | CARD12 (CLAN) interacts with CASP1. This interaction was modelled on a demonstrated interaction between human CARD12 and CASP1 from an unspecified species. | BIND | 15107016 |
| NLRC4 | CARD12 | CLAN | CLAN1 | CLANA | CLANB | CLANC | CLAND | CLR2.1 | IPAF | NLR family, CARD domain containing 4 | - | HPRD | 11390368 |11472070|11472070 |
| NLRP1 | CARD7 | CLR17.1 | DEFCAP | DEFCAP-L/S | DKFZp586O1822 | KIAA0926 | NAC | NALP1 | PP1044 | SLEV1 | VAMAS1 | NLR family, pyrin domain containing 1 | NALP1 (NAC) interacts with CASP1. This interaction was modelled on a demonstrated interaction between human NALP1 and CASP1 from an unspecified species. | BIND | 15107016 |
| NOD1 | CARD4 | CLR7.1 | NLRC1 | nucleotide-binding oligomerization domain containing 1 | - | HPRD,BioGRID | 10329646 |
| NOD2 | ACUG | BLAU | CARD15 | CD | CLR16.3 | IBD1 | NLRC2 | NOD2B | PSORAS1 | nucleotide-binding oligomerization domain containing 2 | Affinity Capture-Western | BioGRID | 15107016 |
| NOD2 | ACUG | BLAU | CARD15 | CD | CLR16.3 | IBD1 | NLRC2 | NOD2B | PSORAS1 | nucleotide-binding oligomerization domain containing 2 | CARD15 (Nod2) interacts with CASP1. This interaction was modelled on a demonstrated interaction between human CARD15 and CASP1 from an unspecified species. | BIND | 15107016 |
| PARK2 | AR-JP | LPRS2 | PDJ | PRKN | Parkinson disease (autosomal recessive, juvenile) 2, parkin | - | HPRD | 12692130 |
| PSEN1 | AD3 | FAD | PS1 | S182 | presenilin 1 | PS-1 interacts with caspase 1. This interaction was modeled on a demonstrated interaction between human PS1 and mouse caspase 1. | BIND | 10069390 |
| PYCARD | ASC | CARD5 | MGC10332 | TMS | TMS-1 | TMS1 | PYD and CARD domain containing | - | HPRD | 11967258 |12019269 |12191486 |
| RIPK2 | CARD3 | CARDIAK | CCK | GIG30 | RICK | RIP2 | receptor-interacting serine-threonine kinase 2 | - | HPRD,BioGRID | 9705938 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG CYTOSOLIC DNA SENSING PATHWAY | 56 | 44 | All SZGR 2.0 genes in this pathway |
| KEGG AMYOTROPHIC LATERAL SCLEROSIS ALS | 53 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CASPASE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA D4GDI PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| PID IL1 PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID P53 DOWNSTREAM PATHWAY | 137 | 94 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| PID ANTHRAX PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME NOD1 2 SIGNALING PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME THE NLRP3 INFLAMMASOME | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME INFLAMMASOMES | 17 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| LIU PROSTATE CANCER DN | 481 | 290 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SCHUETZ BREAST CANCER DUCTAL INVASIVE UP | 351 | 230 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| THUM SYSTOLIC HEART FAILURE UP | 423 | 283 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL DN | 60 | 39 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| GAL LEUKEMIC STEM CELL DN | 244 | 153 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS UP | 269 | 146 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS 8HR DN | 129 | 84 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 48HR UP | 128 | 95 | All SZGR 2.0 genes in this pathway |
| KOKKINAKIS METHIONINE DEPRIVATION 96HR UP | 117 | 84 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 UP | 121 | 71 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 DN | 229 | 142 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| BAKER HEMATOPOESIS STAT1 TARGETS | 10 | 9 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| SCHAVOLT TARGETS OF TP53 AND TP63 | 16 | 12 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| XU HGF TARGETS REPRESSED BY AKT1 DN | 95 | 58 | All SZGR 2.0 genes in this pathway |
| GRUETZMANN PANCREATIC CANCER UP | 358 | 245 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1 | 229 | 137 | All SZGR 2.0 genes in this pathway |
| KHETCHOUMIAN TRIM24 TARGETS UP | 47 | 38 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| GUENTHER GROWTH SPHERICAL VS ADHERENT DN | 26 | 19 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| MORI SMALL PRE BII LYMPHOCYTE DN | 76 | 52 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| ZEILSTRA CD44 TARGETS DN | 7 | 5 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST DN | 309 | 206 | All SZGR 2.0 genes in this pathway |
| SANA RESPONSE TO IFNG UP | 78 | 50 | All SZGR 2.0 genes in this pathway |
| SANA TNF SIGNALING UP | 83 | 56 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 11Q23 DELETION | 23 | 18 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA UP | 64 | 46 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED UP | 183 | 111 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT DN | 102 | 67 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| SCHURINGA STAT5A TARGETS DN | 18 | 10 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| HU GENOTOXIC DAMAGE 24HR | 35 | 22 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR DN | 298 | 200 | All SZGR 2.0 genes in this pathway |
| BROWNE INTERFERON RESPONSIVE GENES | 68 | 44 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| JI RESPONSE TO FSH UP | 74 | 56 | All SZGR 2.0 genes in this pathway |
| BILD HRAS ONCOGENIC SIGNATURE | 261 | 166 | All SZGR 2.0 genes in this pathway |
| BILD SRC ONCOGENIC SIGNATURE | 62 | 38 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION DN | 281 | 179 | All SZGR 2.0 genes in this pathway |
| OUILLETTE CLL 13Q14 DELETION UP | 74 | 40 | All SZGR 2.0 genes in this pathway |
| MARZEC IL2 SIGNALING DN | 36 | 24 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D DN | 270 | 181 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| IZADPANAH STEM CELL ADIPOSE VS BONE UP | 126 | 92 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D DN | 78 | 34 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| ICHIBA GRAFT VERSUS HOST DISEASE 35D UP | 131 | 79 | All SZGR 2.0 genes in this pathway |
| WUNDER INFLAMMATORY RESPONSE AND CHOLESTEROL UP | 58 | 38 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 DN | 315 | 201 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 UP | 408 | 276 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS UP | 395 | 249 | All SZGR 2.0 genes in this pathway |
| HAN SATB1 TARGETS DN | 442 | 275 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS DN | 42 | 23 | All SZGR 2.0 genes in this pathway |
| KUROZUMI RESPONSE TO ONCOCYTIC VIRUS | 44 | 30 | All SZGR 2.0 genes in this pathway |
| GYORFFY DOXORUBICIN RESISTANCE | 56 | 34 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 DN | 69 | 38 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC DN | 253 | 192 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 9 | 76 | 45 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST VS SYNOVIAL SARCOMA DN | 20 | 17 | All SZGR 2.0 genes in this pathway |
| NIELSEN SYNOVIAL SARCOMA DN | 20 | 13 | All SZGR 2.0 genes in this pathway |
| LI INDUCED T TO NATURAL KILLER UP | 307 | 182 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |
| CHYLA CBFA2T3 TARGETS UP | 387 | 225 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE DN | 99 | 74 | All SZGR 2.0 genes in this pathway |