Gene Page: APH1B
Summary ?
| GeneID | 83464 |
| Symbol | APH1B |
| Synonyms | APH-1B|PRO1328|PSFL|TAAV688 |
| Description | aph-1 homolog B, gamma secretase subunit |
| Reference | MIM:607630|HGNC:HGNC:24080|Ensembl:ENSG00000138613|HPRD:06361|Vega:OTTHUMG00000132863 |
| Gene type | protein-coding |
| Map location | 15q22.2 |
| Pascal p-value | 3.584E-4 |
| Sherlock p-value | 0.226 |
| Fetal beta | -0.449 |
| Support | NEUROTROPHIN SIGNALING |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
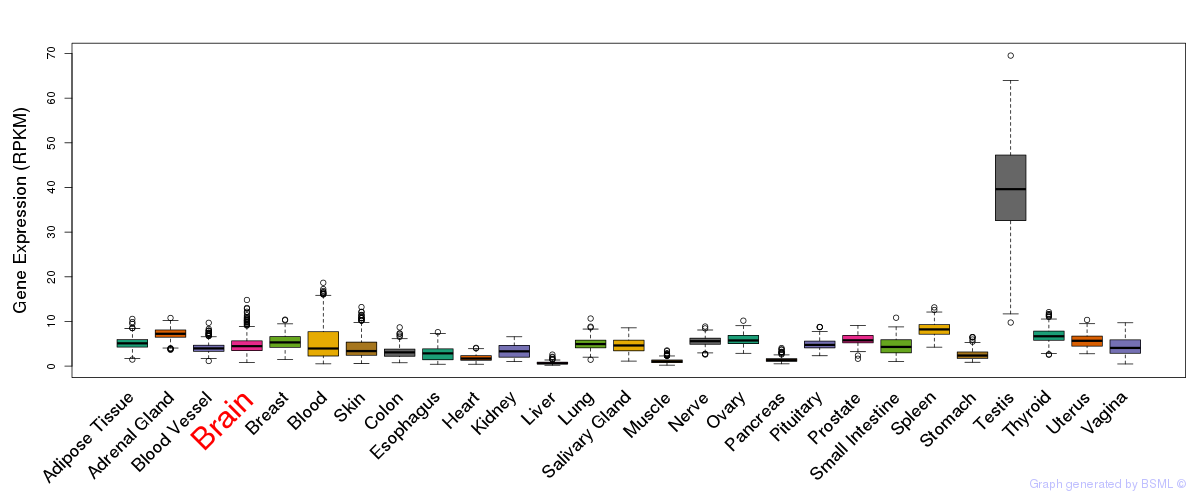
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RBCK1 | 0.81 | 0.80 |
| LMF2 | 0.80 | 0.81 |
| TYK2 | 0.80 | 0.76 |
| SLC12A9 | 0.78 | 0.77 |
| C19orf6 | 0.78 | 0.77 |
| ITFG3 | 0.77 | 0.77 |
| CDK9 | 0.76 | 0.74 |
| C7orf27 | 0.76 | 0.76 |
| D2HGDH | 0.76 | 0.77 |
| AC004410.1 | 0.76 | 0.77 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.53 | -0.56 |
| AF347015.31 | -0.48 | -0.48 |
| CLEC2B | -0.48 | -0.53 |
| SYCP3 | -0.48 | -0.49 |
| GNG11 | -0.47 | -0.49 |
| MT-CO2 | -0.46 | -0.46 |
| AL050337.1 | -0.46 | -0.47 |
| AF347015.27 | -0.45 | -0.47 |
| C1orf54 | -0.45 | -0.45 |
| AF347015.8 | -0.45 | -0.45 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID PS1 PATHWAY | 46 | 39 | All SZGR 2.0 genes in this pathway |
| PID P75 NTR PATHWAY | 69 | 51 | All SZGR 2.0 genes in this pathway |
| PID SYNDECAN 3 PATHWAY | 17 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALLING BY NGF | 217 | 167 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB4 | 90 | 67 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR SIGNALING BY ERBB4 | 38 | 30 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | 27 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH4 | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH2 | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH1 | 70 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH3 | 12 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATED PROTEOLYSIS OF P75NTR | 10 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME NRIF SIGNALS CELL DEATH FROM THE NUCLEUS | 15 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL DEATH SIGNALLING VIA NRAGE NRIF AND NADE | 60 | 43 | All SZGR 2.0 genes in this pathway |
| REACTOME P75 NTR RECEPTOR MEDIATED SIGNALLING | 81 | 61 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY NOTCH | 103 | 64 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| FURUKAWA DUSP6 TARGETS PCI35 UP | 74 | 32 | All SZGR 2.0 genes in this pathway |
| ALCALA APOPTOSIS | 88 | 60 | All SZGR 2.0 genes in this pathway |
| COWLING MYCN TARGETS | 43 | 27 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH MLL FUSIONS | 78 | 45 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA MF DN | 41 | 28 | All SZGR 2.0 genes in this pathway |