Gene Page: RASSF5
Summary ?
| GeneID | 83593 |
| Symbol | RASSF5 |
| Synonyms | Maxp1|NORE1|NORE1A|NORE1B|RAPL|RASSF3 |
| Description | Ras association domain family member 5 |
| Reference | MIM:607020|HGNC:HGNC:17609|Ensembl:ENSG00000266094|HPRD:06117|Vega:OTTHUMG00000184616 |
| Gene type | protein-coding |
| Map location | 1q32.1 |
| TADA p-value | 8.32E-4 |
| Fetal beta | -3.613 |
| DMG | 2 (# studies) |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RASSF5 | chr1 | 206757974..206757975 | CA | C | NM_182663 NM_182664 NM_182664 NM_182665 | . . p.337*>x . | frameshift frameshift read-through frameshift | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg16554516 | 1 | 206680868 | RASSF5 | 4.72E-5 | -0.347 | 0.021 | DMG:Wockner_2014 |
| cg24450312 | 1 | 206681158 | RASSF5 | -0.03 | 0.25 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
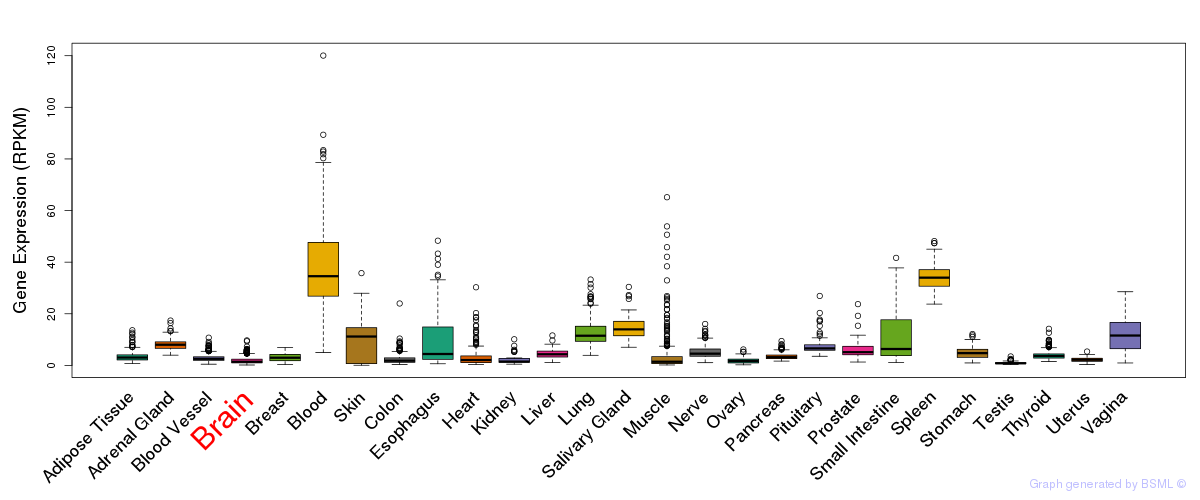
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| HRAS | C-BAS/HAS | C-H-RAS | C-HA-RAS1 | CTLO | H-RASIDX | HAMSV | HRAS1 | K-RAS | N-RAS | RASH1 | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | Reconstituted Complex | BioGRID | 9488663 |
| KRAS | C-K-RAS | K-RAS2A | K-RAS2B | K-RAS4A | K-RAS4B | KI-RAS | KRAS1 | KRAS2 | NS3 | RASK2 | v-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog | - | HPRD,BioGRID | 11857081 |
| MRAS | FLJ42964 | M-RAs | R-RAS3 | RRAS3 | muscle RAS oncogene homolog | - | HPRD,BioGRID | 11857081 |
| RAP1B | DKFZp586H0723 | K-REV | RAL1B | RAP1B, member of RAS oncogene family | - | HPRD | 9488663 |
| RAP2A | K-REV | KREV | RAP2 | RbBP-30 | RAP2A, member of RAS oncogene family | - | HPRD,BioGRID | 11857081 |
| RASSF1 | 123F2 | NORE2A | RASSF1A | RDA32 | REH3P21 | Ras association (RalGDS/AF-6) domain family member 1 | - | HPRD,BioGRID | 11857081 |
| RASSF2 | DKFZp781O1747 | KIAA0168 | RASFADIN | Ras association (RalGDS/AF-6) domain family member 2 | Reconstituted Complex | BioGRID | 11857081 |
| RASSF5 | MGC10823 | MGC17344 | Maxp1 | NORE1 | NORE1A | NORE1B | RAPL | RASSF3 | Ras association (RalGDS/AF-6) domain family member 5 | Reconstituted Complex | BioGRID | 11857081 |
| RRAS | - | related RAS viral (r-ras) oncogene homolog | - | HPRD,BioGRID | 11857081 |
| RRAS2 | TC21 | related RAS viral (r-ras) oncogene homolog 2 | - | HPRD | 11857081 |
| STK4 | DKFZp686A2068 | KRS2 | MST1 | YSK3 | serine/threonine kinase 4 | - | HPRD | 11864565 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LEUKOCYTE TRANSENDOTHELIAL MIGRATION | 118 | 78 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG NON SMALL CELL LUNG CANCER | 54 | 47 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC DN | 537 | 339 | All SZGR 2.0 genes in this pathway |
| GOZGIT ESR1 TARGETS DN | 781 | 465 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP | 612 | 367 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 AND TP63 TARGETS | 205 | 145 | All SZGR 2.0 genes in this pathway |
| SCHLESINGER METHYLATED DE NOVO IN CANCER | 88 | 64 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS DN | 261 | 155 | All SZGR 2.0 genes in this pathway |
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 1Q32 AMPLICON | 13 | 5 | All SZGR 2.0 genes in this pathway |
| ZHAN MULTIPLE MYELOMA CD1 AND CD2 DN | 51 | 32 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS DN | 32 | 25 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 9 | 92 | 59 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE UP | 578 | 341 | All SZGR 2.0 genes in this pathway |
| BONCI TARGETS OF MIR15A AND MIR16 1 | 91 | 75 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| CHEN METABOLIC SYNDROM NETWORK | 1210 | 725 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME3 AND H3K27ME3 | 142 | 103 | All SZGR 2.0 genes in this pathway |
| COULOUARN TEMPORAL TGFB1 SIGNATURE DN | 138 | 99 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K4ME3 AND H3K27ME3 | 210 | 148 | All SZGR 2.0 genes in this pathway |
| LU EZH2 TARGETS DN | 414 | 237 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE EARLY LATE | 317 | 190 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |