Gene Page: STK40
Summary ?
| GeneID | 83931 |
| Symbol | STK40 |
| Synonyms | SHIK|SgK495 |
| Description | serine/threonine kinase 40 |
| Reference | MIM:609437|HGNC:HGNC:21373|Ensembl:ENSG00000196182|HPRD:11359|Vega:OTTHUMG00000008238 |
| Gene type | protein-coding |
| Map location | 1p34.3 |
| Pascal p-value | 0.589 |
| Sherlock p-value | 0.378 |
| Fetal beta | 0.501 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 1 |
| Expression | Meta-analysis of gene expression | P value: 1.836 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25722983 | 1 | 36840028 | STK40 | 5.27E-5 | -0.007 | 0.098 | DMG:Montano_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs16858695 | chr1 | 162193737 | STK40 | 83931 | 0.2 | trans | ||
| rs2014805 | chr2 | 175165192 | STK40 | 83931 | 0.06 | trans | ||
| rs7593761 | chr2 | 197275602 | STK40 | 83931 | 0.17 | trans | ||
| rs16827037 | chr3 | 117203284 | STK40 | 83931 | 0.13 | trans | ||
| rs7664027 | chr4 | 169121200 | STK40 | 83931 | 0.01 | trans | ||
| rs17053782 | chr4 | 169127022 | STK40 | 83931 | 0.01 | trans | ||
| rs11722969 | chr4 | 169133757 | STK40 | 83931 | 0.01 | trans | ||
| rs6846811 | chr4 | 169234535 | STK40 | 83931 | 0.14 | trans | ||
| rs904451 | chr4 | 186630945 | STK40 | 83931 | 0.08 | trans | ||
| rs887719 | chr7 | 21184272 | STK40 | 83931 | 0.07 | trans | ||
| rs320082 | chr7 | 29105404 | STK40 | 83931 | 0 | trans | ||
| rs317720 | chr7 | 29110633 | STK40 | 83931 | 0.08 | trans | ||
| rs317714 | chr7 | 29115553 | STK40 | 83931 | 1.473E-5 | trans | ||
| rs317724 | chr7 | 29120565 | STK40 | 83931 | 0.08 | trans | ||
| rs7850851 | chr9 | 37928770 | STK40 | 83931 | 0.15 | trans | ||
| rs10834670 | chr11 | 25404015 | STK40 | 83931 | 0.14 | trans | ||
| rs10834671 | chr11 | 25404269 | STK40 | 83931 | 0.11 | trans | ||
| rs4497566 | chr13 | 51694324 | STK40 | 83931 | 8.11E-4 | trans | ||
| rs1307687 | 0 | STK40 | 83931 | 0.08 | trans |
Section II. Transcriptome annotation
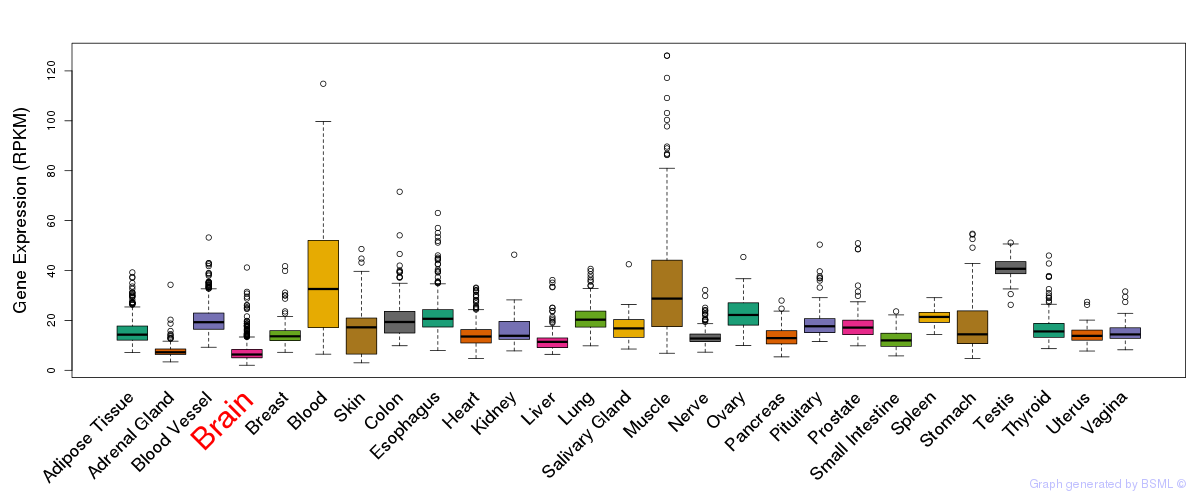
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005730 | nucleolus | IDA | 18029348 | |
| GO:0005737 | cytoplasm | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENESE HDAC2 TARGETS DN | 133 | 77 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| GALINDO IMMUNE RESPONSE TO ENTEROTOXIN | 85 | 67 | All SZGR 2.0 genes in this pathway |
| SARTIPY NORMAL AT INSULIN RESISTANCE UP | 34 | 27 | All SZGR 2.0 genes in this pathway |
| YAMANAKA GLIOBLASTOMA SURVIVAL UP | 11 | 7 | All SZGR 2.0 genes in this pathway |
| YOSHIOKA LIVER CANCER EARLY RECURRENCE DN | 65 | 31 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| KUMAR PATHOGEN LOAD BY MACROPHAGES | 275 | 155 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 259 | 265 | m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-129-5p | 2045 | 2051 | 1A | hsa-miR-129brain | CUUUUUGCGGUCUGGGCUUGC |
| hsa-miR-129-5p | CUUUUUGCGGUCUGGGCUUGCU | ||||
| miR-134 | 2040 | 2046 | 1A | hsa-miR-134brain | UGUGACUGGUUGACCAGAGGG |
| miR-137 | 2103 | 2109 | 1A | hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG |
| hsa-miR-137 | UAUUGCUUAAGAAUACGCGUAG | ||||
| miR-21 | 387 | 393 | m8 | hsa-miR-21brain | UAGCUUAUCAGACUGAUGUUGA |
| hsa-miR-590 | GAGCUUAUUCAUAAAAGUGCAG | ||||
| miR-31 | 1475 | 1482 | 1A,m8 | hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG |
| hsa-miR-31 | AGGCAAGAUGCUGGCAUAGCUG | ||||
| miR-381 | 2054 | 2060 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.