Gene Page: CASP8
Summary ?
| GeneID | 841 |
| Symbol | CASP8 |
| Synonyms | ALPS2B|CAP4|Casp-8|FLICE|MACH|MCH5 |
| Description | caspase 8 |
| Reference | MIM:601763|HGNC:HGNC:1509|Ensembl:ENSG00000064012|HPRD:03459|Vega:OTTHUMG00000132821 |
| Gene type | protein-coding |
| Map location | 2q33-q34 |
| Pascal p-value | 0.033 |
| Fetal beta | 0.114 |
| eGene | Anterior cingulate cortex BA24 Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs59308963 | 2 | 202123479 | CASP8 | ENSG00000064012.17 | 3.101E-8 | 0 | 25313 | gtex_brain_ba24 |
| rs55971105 | 2 | 202123717 | CASP8 | ENSG00000064012.17 | 3.137E-6 | 0 | 25551 | gtex_brain_ba24 |
| rs6735656 | 2 | 202124502 | CASP8 | ENSG00000064012.17 | 3.32E-8 | 0 | 26336 | gtex_brain_ba24 |
| rs6754084 | 2 | 202124997 | CASP8 | ENSG00000064012.17 | 3.111E-8 | 0 | 26831 | gtex_brain_ba24 |
| rs1861270 | 2 | 202126615 | CASP8 | ENSG00000064012.17 | 3.111E-8 | 0 | 28449 | gtex_brain_ba24 |
| rs6435074 | 2 | 202127947 | CASP8 | ENSG00000064012.17 | 3.105E-8 | 0 | 29781 | gtex_brain_ba24 |
| rs2349070 | 2 | 202130308 | CASP8 | ENSG00000064012.17 | 3.099E-8 | 0 | 32142 | gtex_brain_ba24 |
| rs10931936 | 2 | 202143928 | CASP8 | ENSG00000064012.17 | 8.612E-8 | 0 | 45762 | gtex_brain_ba24 |
| rs3769818 | 2 | 202151163 | CASP8 | ENSG00000064012.17 | 8.612E-8 | 0 | 52997 | gtex_brain_ba24 |
| rs700635 | 2 | 202153225 | CASP8 | ENSG00000064012.17 | 8.612E-8 | 0 | 55059 | gtex_brain_ba24 |
| rs6714430 | 2 | 202153684 | CASP8 | ENSG00000064012.17 | 8.612E-8 | 0 | 55518 | gtex_brain_ba24 |
| rs6743068 | 2 | 202153920 | CASP8 | ENSG00000064012.17 | 8.612E-8 | 0 | 55754 | gtex_brain_ba24 |
| rs10200279 | 2 | 202170655 | CASP8 | ENSG00000064012.17 | 8.713E-8 | 0 | 72489 | gtex_brain_ba24 |
| rs6719014 | 2 | 202176024 | CASP8 | ENSG00000064012.17 | 8.982E-8 | 0 | 77858 | gtex_brain_ba24 |
| rs7582362 | 2 | 202176294 | CASP8 | ENSG00000064012.17 | 8.99E-8 | 0 | 78128 | gtex_brain_ba24 |
| rs6731171 | 2 | 202179011 | CASP8 | ENSG00000064012.17 | 1.381E-6 | 0 | 80845 | gtex_brain_ba24 |
| rs1830298 | 2 | 202181247 | CASP8 | ENSG00000064012.17 | 2.584E-7 | 0 | 83081 | gtex_brain_ba24 |
| rs9677180 | 2 | 202184331 | CASP8 | ENSG00000064012.17 | 9.479E-8 | 0 | 86165 | gtex_brain_ba24 |
| rs2349073 | 2 | 202186986 | CASP8 | ENSG00000064012.17 | 1.36E-7 | 0 | 88820 | gtex_brain_ba24 |
| rs10197246 | 2 | 202204741 | CASP8 | ENSG00000064012.17 | 6.761E-7 | 0 | 106575 | gtex_brain_ba24 |
| rs2110687 | 2 | 202223403 | CASP8 | ENSG00000064012.17 | 2.188E-6 | 0 | 125237 | gtex_brain_ba24 |
Section II. Transcriptome annotation
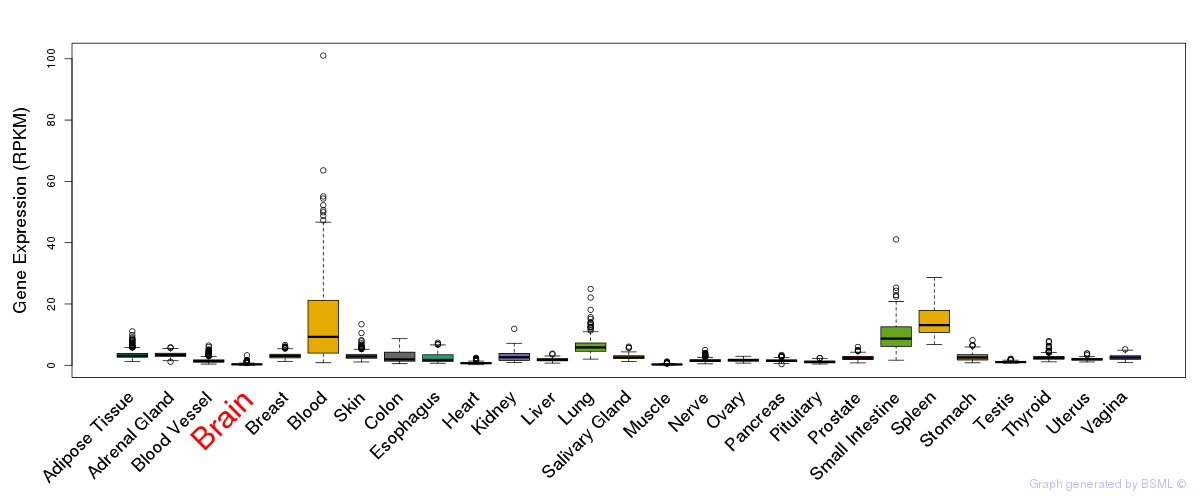
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NEXN | 0.87 | 0.51 |
| CARD11 | 0.86 | 0.58 |
| FAM19A4 | 0.84 | 0.64 |
| SUSD2 | 0.83 | 0.75 |
| SMPX | 0.83 | 0.52 |
| DPYS | 0.82 | 0.45 |
| RGS16 | 0.81 | 0.18 |
| AL133445.2 | 0.80 | 0.34 |
| FAM20A | 0.80 | 0.02 |
| ABHD12B | 0.79 | 0.71 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PLEKHO1 | -0.34 | -0.67 |
| NR2C2AP | -0.32 | -0.67 |
| SHF | -0.30 | -0.62 |
| MPP3 | -0.30 | -0.60 |
| C19orf57 | -0.30 | -0.59 |
| TUBB2B | -0.29 | -0.58 |
| SAMD14 | -0.29 | -0.62 |
| KIAA1949 | -0.29 | -0.56 |
| MARCKSL1 | -0.29 | -0.59 |
| STAG3 | -0.29 | -0.57 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| APAF1 | CED4 | DKFZp781B1145 | apoptotic peptidase activating factor 1 | - | HPRD,BioGRID | 9539746 |
| APP | AAA | ABETA | ABPP | AD1 | APPI | CTFgamma | CVAP | PN2 | amyloid beta (A4) precursor protein | - | HPRD | 10911620 |
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | Androgen Receptor interacts with caspase 8. | BIND | 9535906 |
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Co-purification | BioGRID | 15659383 |
| ATN1 | B37 | D12S755E | DRPLA | NOD | atrophin 1 | Atrophin-1 interacts with caspase 8. | BIND | 9535906 |
| BCAP31 | 6C6-AG | BAP31 | CDM | DXS1357E | B-cell receptor-associated protein 31 | - | HPRD,BioGRID | 9334338 |
| BCL2 | Bcl-2 | B-cell CLL/lymphoma 2 | - | HPRD,BioGRID | 11406564 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | - | HPRD | 11733517 |
| BCL2L1 | BCL-XL/S | BCL2L | BCLX | Bcl-X | DKFZp781P2092 | bcl-xL | bcl-xS | BCL2-like 1 | Phenotypic Suppression | BioGRID | 11832478 |
| BFAR | BAR | RNF47 | bifunctional apoptosis regulator | - | HPRD | 10716992 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Biochemical Activity Co-purification | BioGRID | 11832478 |15659383 |
| BIRC2 | API1 | HIAP2 | Hiap-2 | MIHB | RNF48 | cIAP1 | baculoviral IAP repeat-containing 2 | Affinity Capture-Western | BioGRID | 12887920 |
| CASP1 | ICE | IL1BC | P45 | caspase 1, apoptosis-related cysteine peptidase (interleukin 1, beta, convertase) | Biochemical Activity | BioGRID | 8962078 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | Affinity Capture-Western Biochemical Activity Co-purification | BioGRID | 8962078 |11832478 |12887920 |15659383 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | - | HPRD | 9325248 |
| CASP14 | MGC119078 | MGC119079 | caspase 14, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 9792675 |
| CASP2 | CASP-2 | ICH-1L | ICH-1L/1S | ICH1 | NEDD2 | caspase 2, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 8962078 |11832478 |
| CASP3 | CPP32 | CPP32B | SCA-1 | caspase 3, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 8962078 |11832478 |
| CASP4 | ICE(rel)II | ICEREL-II | ICH-2 | Mih1/TX | TX | caspase 4, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 8962078 |
| CASP6 | MCH2 | caspase 6, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 12232792 |
| CASP7 | CMH-1 | ICE-LAP3 | MCH3 | caspase 7, apoptosis-related cysteine peptidase | Biochemical Activity | BioGRID | 8962078 |11832478 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 10964557 |
| CASP8AP2 | CED-4 | FLASH | FLJ11208 | KIAA1315 | RIP25 | caspase 8 associated protein 2 | - | HPRD,BioGRID | 10235259 |11340079 |
| CASP9 | APAF-3 | APAF3 | CASPASE-9c | ICE-LAP6 | MCH6 | caspase 9, apoptosis-related cysteine peptidase | Biochemical Activity Phenotypic Suppression | BioGRID | 8962078 |11832478 |
| CDC2L2 | CDC2L3 | CDK11-p110 | CDK11-p46 | CDK11-p58 | MGC131975 | PITSLRE | p58GTA | cell division cycle 2-like 2 (PITSLRE proteins) | - | HPRD | 9632733 |
| CDH1 | Arc-1 | CD324 | CDHE | ECAD | LCAM | UVO | cadherin 1, type 1, E-cadherin (epithelial) | - | HPRD | 12189238 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | MRIT-beta-1 interacts with FLICE. This interaction was modelled on a demonstrated interaction between human MRIT-beta-1 and FLICE from an unspecified species. | BIND | 9326610 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9208847 |9228018 |9289491 |9326610 |11741985 |12215447 |12887920 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | - | HPRD | 9208847 |9289491 |12215447 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | MRIT-alpha-1 interacts with FLICE. This interaction was modelled on a demonstrated interaction between human MRIT-alpha-1 and FLICE from an unspecified species. | BIND | 9326610 |
| CHD3 | Mi-2a | Mi2-ALPHA | ZFH | chromodomain helicase DNA binding protein 3 | Two-hybrid | BioGRID | 16169070 |
| CHUK | IKBKA | IKK-alpha | IKK1 | IKKA | NFKBIKA | TCF16 | conserved helix-loop-helix ubiquitous kinase | - | HPRD,BioGRID | 11002417 |
| CRADD | MGC9163 | RAIDD | CASP2 and RIPK1 domain containing adaptor with death domain | - | HPRD | 11002417 |
| CTNNB1 | CTNNB | DKFZp686D02253 | FLJ25606 | FLJ37923 | catenin (cadherin-associated protein), beta 1, 88kDa | - | HPRD | 12189238 |
| DEDD | CASP8IP1 | DEDD1 | DEFT | FLDED1 | KE05 | death effector domain containing | - | HPRD,BioGRID | 12527898 |
| DEDD2 | FLAME-3 | death effector domain containing 2 | - | HPRD,BioGRID | 11965497 |12527898|12527898 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Co-purification | BioGRID | 15659383 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | CASP8 (caspase-8) interacts with FADD. | BIND | 15782135 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD,BioGRID | 12107169 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | - | HPRD | 11717445 |12107169|12107169 |
| FADD | GIG3 | MGC8528 | MORT1 | Fas (TNFRSF6)-associated via death domain | caspase-8 interacts with FADD. | BIND | 12220669 |
| FAF1 | CGI-03 | FLJ37524 | HFAF1s | UBXD12 | UBXN3A | hFAF1 | Fas (TNFRSF6) associated factor 1 | - | HPRD | 12702723 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD | 11101867 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Affinity Capture-Western Co-purification | BioGRID | 9208847 |9325248 |15659383 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | Affinity Capture-Western Co-purification | BioGRID | 12887920 |15659383 |
| HIP1 | ILWEQ | MGC126506 | huntingtin interacting protein 1 | - | HPRD,BioGRID | 11788820 |
| HTT | HD | IT15 | huntingtin | Htt interacts with caspase 8. | BIND | 14713958 |
| IFT57 | ESRRBL1 | FLJ10147 | HIPPI | MHS4R2 | intraflagellar transport 57 homolog (Chlamydomonas) | - | HPRD,BioGRID | 11788820 |
| IKBKB | FLJ40509 | IKK-beta | IKK2 | IKKB | MGC131801 | NFKBIKB | inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta | - | HPRD,BioGRID | 11002417 |
| MAP3K14 | FTDCR1B | HS | HSNIK | NIK | mitogen-activated protein kinase kinase kinase 14 | - | HPRD,BioGRID | 11002417 |
| MAP4K4 | FLH21957 | FLJ10410 | FLJ20373 | FLJ90111 | HGK | KIAA0687 | NIK | mitogen-activated protein kinase kinase kinase kinase 4 | - | HPRD | 11002417 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Co-purification | BioGRID | 15659383 |
| MSN | - | moesin | Co-purification | BioGRID | 15659383 |
| NOD1 | CARD4 | CLR7.1 | NLRC1 | nucleotide-binding oligomerization domain containing 1 | - | HPRD,BioGRID | 10329646 |
| NOL3 | ARC | CARD2 | MYC | MYP | NOP | NOP30 | nucleolar protein 3 (apoptosis repressor with CARD domain) | ARC interacts with procaspase-8. | BIND | 15383280 |
| NOL3 | ARC | CARD2 | MYC | MYP | NOP | NOP30 | nucleolar protein 3 (apoptosis repressor with CARD domain) | - | HPRD,BioGRID | 9560245 |
| NR1H4 | BAR | FXR | HRR-1 | HRR1 | MGC163445 | RIP14 | nuclear receptor subfamily 1, group H, member 4 | - | HPRD,BioGRID | 11733517 |
| PEA15 | HMAT1 | HUMMAT1H | MAT1 | MAT1H | PEA-15 | PED | phosphoprotein enriched in astrocytes 15 | - | HPRD,BioGRID | 10442631 |10493725 |
| PIAS1 | DDXBP1 | GBP | GU/RH-II | MGC141878 | MGC141879 | ZMIZ3 | protein inhibitor of activated STAT, 1 | CASP8 (caspase-8) interacts with PIAS-1. | BIND | 15782135 |
| PICALM | CALM | CLTH | LAP | phosphatidylinositol binding clathrin assembly protein | - | HPRD | 10926122 |
| RALBP1 | RIP1 | RLIP1 | RLIP76 | ralA binding protein 1 | Affinity Capture-Western | BioGRID | 12887920 |
| RB1 | OSRC | RB | p105-Rb | pRb | pp110 | retinoblastoma 1 | CASP8 (caspase-8) cleaves RB-1 (hRB) into the p68 form and to a lesser extent the p61 form. | BIND | 15735701 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Co-purification | BioGRID | 15659383 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD,BioGRID | 11002417 |
| RIPK2 | CARD3 | CARDIAK | CCK | GIG30 | RICK | RIP2 | receptor-interacting serine-threonine kinase 2 | - | HPRD | 9575181 |
| RYBP | AAP1 | DEDAF | YEAF1 | RING1 and YY1 binding protein | - | HPRD | 11395500 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | CASP8 (caspase-8) interacts with SUMO1. | BIND | 15782135 |
| TFCP2 | CP2 | LBP-1C | LSF | SEF | TFCP2C | transcription factor CP2 | Two-hybrid | BioGRID | 16169070 |
| TNFRSF10A | APO2 | CD261 | DR4 | MGC9365 | TRAILR-1 | TRAILR1 | tumor necrosis factor receptor superfamily, member 10a | Affinity Capture-Western | BioGRID | 9325248 |
| TNFRSF10A | APO2 | CD261 | DR4 | MGC9365 | TRAILR-1 | TRAILR1 | tumor necrosis factor receptor superfamily, member 10a | - | HPRD | 12218097 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | Affinity Capture-Western Co-purification | BioGRID | 9325248 |15659383 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | - | HPRD | 12218097 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Co-purification | BioGRID | 15659383 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD,BioGRID | 12887920 |
| TRAF1 | EBI6 | MGC:10353 | TNF receptor-associated factor 1 | Affinity Capture-Western Biochemical Activity | BioGRID | 11098060 |12887920 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Affinity Capture-Western | BioGRID | 12887920 |
| VIM | FLJ36605 | vimentin | Two-hybrid | BioGRID | 16169070 |
| XIAP | API3 | BIRC4 | ILP1 | MIHA | XLP2 | X-linked inhibitor of apoptosis | Phenotypic Suppression | BioGRID | 11832478 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG NOD LIKE RECEPTOR SIGNALING PATHWAY | 62 | 47 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG HUNTINGTONS DISEASE | 185 | 109 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG VIRAL MYOCARDITIS | 73 | 58 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CASPASE PATHWAY | 23 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CERAMIDE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA D4GDI PATHWAY | 13 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MITOCHONDRIA PATHWAY | 21 | 15 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SODD PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR1 PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| ST TUMOR NECROSIS FACTOR PATHWAY | 29 | 20 | All SZGR 2.0 genes in this pathway |
| SA CASPASE CASCADE | 19 | 13 | All SZGR 2.0 genes in this pathway |
| SA FAS SIGNALING | 9 | 5 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID AR PATHWAY | 61 | 46 | All SZGR 2.0 genes in this pathway |
| PID TRAIL PATHWAY | 28 | 20 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| PID CASPASE PATHWAY | 52 | 39 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC CLEAVAGE OF CELLULAR PROTEINS | 40 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | 13 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME NOD1 2 SIGNALING PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEOTIDE BINDING DOMAIN LEUCINE RICH REPEAT CONTAINING RECEPTOR NLR SIGNALING PATHWAYS | 46 | 33 | All SZGR 2.0 genes in this pathway |
| REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | 30 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOTIC EXECUTION PHASE | 54 | 37 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLUE DN | 61 | 35 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| MARKS HDAC TARGETS UP | 23 | 15 | All SZGR 2.0 genes in this pathway |
| DUTTA APOPTOSIS VIA NFKB | 33 | 25 | All SZGR 2.0 genes in this pathway |
| HUMMERICH SKIN CANCER PROGRESSION UP | 88 | 58 | All SZGR 2.0 genes in this pathway |
| RASHI RESPONSE TO IONIZING RADIATION 5 | 147 | 89 | All SZGR 2.0 genes in this pathway |
| SCHEIDEREIT IKK INTERACTING PROTEINS | 58 | 45 | All SZGR 2.0 genes in this pathway |
| LA MEN1 TARGETS | 24 | 15 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 1 DN | 169 | 102 | All SZGR 2.0 genes in this pathway |
| SHEPARD CRUSH AND BURN MUTANT UP | 197 | 110 | All SZGR 2.0 genes in this pathway |
| DER IFN ALPHA RESPONSE UP | 74 | 48 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP | 205 | 126 | All SZGR 2.0 genes in this pathway |
| DER IFN BETA RESPONSE UP | 102 | 67 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| DER IFN GAMMA RESPONSE UP | 71 | 45 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| CHIARETTI ACUTE LYMPHOBLASTIC LEUKEMIA ZAP70 | 67 | 33 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| GERHOLD ADIPOGENESIS DN | 64 | 44 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D7 | 40 | 21 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV NHEK DN | 318 | 220 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| DAZARD UV RESPONSE CLUSTER G6 | 153 | 112 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS DN | 314 | 188 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR UP | 783 | 442 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| GALI TP53 TARGETS APOPTOTIC UP | 8 | 7 | All SZGR 2.0 genes in this pathway |
| MOOTHA HUMAN MITODB 6 2002 | 429 | 260 | All SZGR 2.0 genes in this pathway |
| MOOTHA MITOCHONDRIA | 447 | 277 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN PLURIPOTENT STATE DN | 8 | 7 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3 UNMETHYLATED | 536 | 296 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 12 | 79 | 54 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA MESENCHYMAL | 216 | 130 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 36HR | 152 | 88 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |