Gene Page: NR0B2
Summary ?
| GeneID | 8431 |
| Symbol | NR0B2 |
| Synonyms | SHP|SHP1 |
| Description | nuclear receptor subfamily 0 group B member 2 |
| Reference | MIM:604630|HGNC:HGNC:7961|Ensembl:ENSG00000131910|HPRD:05219|Vega:OTTHUMG00000004231 |
| Gene type | protein-coding |
| Map location | 1p36.1 |
| Pascal p-value | 0.005 |
| Fetal beta | 0.068 |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| NR0B2 | chr1 | 27240176 | G | A | NM_021969 | p.86R>W | missense | Schizophrenia | DNM:Fromer_2014 |
Section II. Transcriptome annotation
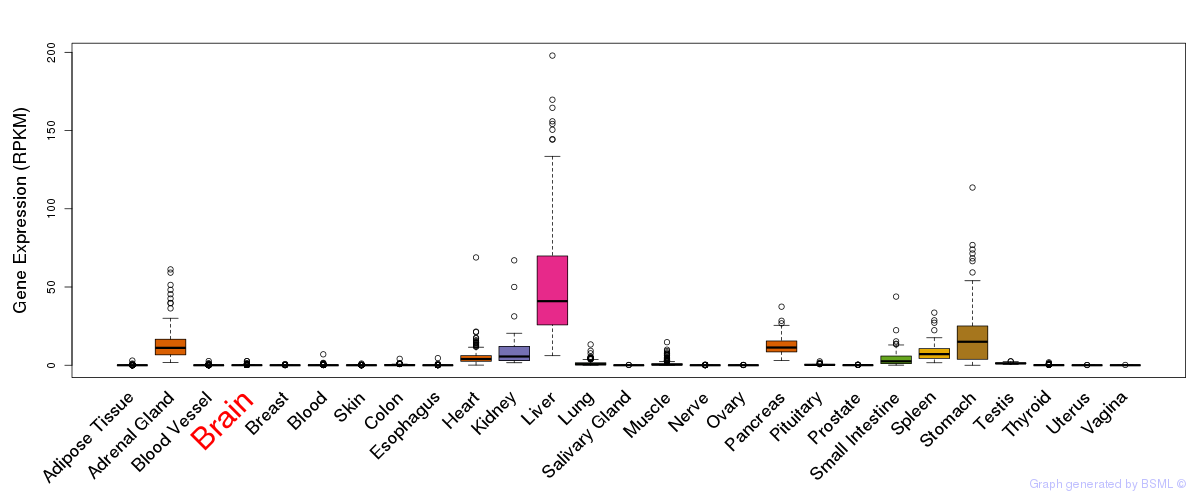
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ADAM11 | 0.88 | 0.88 |
| HISPPD2A | 0.88 | 0.87 |
| AC005277.1 | 0.87 | 0.89 |
| RNF157 | 0.87 | 0.89 |
| TOM1L2 | 0.87 | 0.85 |
| SYNGR1 | 0.87 | 0.87 |
| PCNXL2 | 0.87 | 0.88 |
| SNPH | 0.87 | 0.85 |
| IQSEC3 | 0.87 | 0.87 |
| STXBP1 | 0.87 | 0.86 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.46 | -0.53 |
| C9orf46 | -0.45 | -0.43 |
| GNG11 | -0.44 | -0.46 |
| MYL12A | -0.43 | -0.52 |
| RPL35 | -0.43 | -0.48 |
| CLEC3B | -0.43 | -0.54 |
| RPS20 | -0.43 | -0.50 |
| RPL31 | -0.42 | -0.43 |
| RAB13 | -0.42 | -0.47 |
| RAMP2 | -0.41 | -0.47 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AR | AIS | DHTR | HUMARA | KD | NR3C4 | SBMA | SMAX1 | TFM | androgen receptor | - | HPRD,BioGRID | 11735420 |
| ESR1 | DKFZp686N23123 | ER | ESR | ESRA | Era | NR3A1 | estrogen receptor 1 | - | HPRD,BioGRID | 9773978 |11861507 |
| ESR2 | ER-BETA | ESR-BETA | ESRB | ESTRB | Erb | NR3A2 | estrogen receptor 2 (ER beta) | - | HPRD,BioGRID | 9773978 |10648597 |
| HNF4A | FLJ39654 | HNF4 | HNF4a7 | HNF4a8 | HNF4a9 | MODY | MODY1 | NR2A1 | NR2A21 | TCF | TCF14 | hepatocyte nuclear factor 4, alpha | Reconstituted Complex Two-hybrid | BioGRID | 10594021 |
| NCOA3 | ACTR | AIB-1 | AIB1 | CAGH16 | CTG26 | KAT13B | MGC141848 | RAC3 | SRC3 | TNRC14 | TNRC16 | TRAM-1 | pCIP | nuclear receptor coactivator 3 | Two-hybrid | BioGRID | 11668176 |
| NR1H2 | LXR-b | LXRB | NER | NER-I | RIP15 | UNR | nuclear receptor subfamily 1, group H, member 2 | - | HPRD,BioGRID | 12198243 |
| NR1H3 | LXR-a | LXRA | RLD-1 | nuclear receptor subfamily 1, group H, member 3 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12198243 |
| NR1I3 | CAR | CAR1 | MB67 | MGC150433 | MGC97144 | MGC97209 | nuclear receptor subfamily 1, group I, member 3 | - | HPRD | 8650544|9773978 |
| NR5A2 | B1F | B1F2 | CPF | FTF | FTZ-F1 | FTZ-F1beta | LRH-1 | hB1F | hB1F-2 | nuclear receptor subfamily 5, group A, member 2 | - | HPRD,BioGRID | 11668176 |
| NR5A2 | B1F | B1F2 | CPF | FTF | FTZ-F1 | FTZ-F1beta | LRH-1 | hB1F | hB1F-2 | nuclear receptor subfamily 5, group A, member 2 | hLRH-1 interacts with hSHP. This interaction was modelled on a demonstrated interaction between mouse LRH-1 and human SHP. | BIND | 15707893 |
| POLR2A | MGC75453 | POLR2 | POLRA | RPB1 | RPBh1 | RPO2 | RPOL2 | RpIILS | hRPB220 | hsRPB1 | polymerase (RNA) II (DNA directed) polypeptide A, 220kDa | Reconstituted Complex | BioGRID | 12198243 |
| PPARG | NR1C3 | PPARG1 | PPARG2 | PPARgamma | peroxisome proliferator-activated receptor gamma | - | HPRD,BioGRID | 11696534 |
| RARA | NR1B1 | RAR | retinoic acid receptor, alpha | - | HPRD,BioGRID | 9773978 |
| RXRA | FLJ00280 | FLJ00318 | FLJ16020 | FLJ16733 | MGC102720 | NR2B1 | retinoid X receptor, alpha | - | HPRD,BioGRID | 10594021 |
| RXRB | DAUDI6 | H-2RIIBP | MGC1831 | NR2B2 | RCoR-1 | retinoid X receptor, beta | - | HPRD | 10594021 |
| RXRG | NR2B3 | RXRC | retinoid X receptor, gamma | Reconstituted Complex | BioGRID | 8650544 |
| THRA | AR7 | EAR7 | ERB-T-1 | ERBA | ERBA1 | MGC000261 | MGC43240 | NR1A1 | THRA1 | THRA2 | c-ERBA-1 | thyroid hormone receptor, alpha (erythroblastic leukemia viral (v-erb-a) oncogene homolog, avian) | Reconstituted Complex | BioGRID | 8650544 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BIOCARTA PPARA PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NUCLEARRS PATHWAY | 15 | 12 | All SZGR 2.0 genes in this pathway |
| SIG PIP3 SIGNALING IN B LYMPHOCYTES | 36 | 31 | All SZGR 2.0 genes in this pathway |
| SIG BCR SIGNALING PATHWAY | 46 | 38 | All SZGR 2.0 genes in this pathway |
| ST INTERLEUKIN 4 PATHWAY | 26 | 18 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID ERB GENOMIC PATHWAY | 15 | 8 | All SZGR 2.0 genes in this pathway |
| PID ERA GENOMIC PATHWAY | 65 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | 49 | 36 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| WANG BARRETTS ESOPHAGUS AND ESOPHAGUS CANCER UP | 26 | 12 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION HEMGN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO DN | 200 | 112 | All SZGR 2.0 genes in this pathway |
| HSIAO LIVER SPECIFIC GENES | 244 | 154 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF LCP WITH H3K27ME3 | 70 | 35 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN IPS LCP WITH H3K4ME3 | 174 | 100 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES LCP WITH H3K4ME3 | 142 | 80 | All SZGR 2.0 genes in this pathway |
| SERVITJA LIVER HNF1A TARGETS DN | 157 | 105 | All SZGR 2.0 genes in this pathway |