Gene Page: C7orf50
Summary ?
| GeneID | 84310 |
| Symbol | C7orf50 |
| Synonyms | YCR016W |
| Description | chromosome 7 open reading frame 50 |
| Reference | HGNC:HGNC:22421|Ensembl:ENSG00000146540|HPRD:14413|Vega:OTTHUMG00000151477 |
| Gene type | protein-coding |
| Map location | 7p22.3 |
| Pascal p-value | 0.864 |
| Sherlock p-value | 0.213 |
| Fetal beta | -0.587 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14525935 | 7 | 1105730 | C7orf50 | 2.45E-4 | -0.425 | 0.037 | DMG:Wockner_2014 |
| cg16190225 | 7 | 1163456 | C7orf50 | 4.452E-4 | -0.316 | 0.045 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs113145978 | 7 | 1000578 | C7orf50 | ENSG00000146540.10 | 4.223E-9 | 0 | 177318 | gtex_brain_ba24 |
| rs73257955 | 7 | 1007014 | C7orf50 | ENSG00000146540.10 | 4.617E-9 | 0 | 170882 | gtex_brain_ba24 |
| rs78461900 | 7 | 1007033 | C7orf50 | ENSG00000146540.10 | 4.617E-9 | 0 | 170863 | gtex_brain_ba24 |
| rs6943665 | 7 | 1010909 | C7orf50 | ENSG00000146540.10 | 1.513E-10 | 0 | 166987 | gtex_brain_ba24 |
| rs6462277 | 7 | 1011968 | C7orf50 | ENSG00000146540.10 | 2.621E-10 | 0 | 165928 | gtex_brain_ba24 |
| rs56242247 | 7 | 1012948 | C7orf50 | ENSG00000146540.10 | 2.621E-10 | 0 | 164948 | gtex_brain_ba24 |
| rs55729719 | 7 | 1013180 | C7orf50 | ENSG00000146540.10 | 2.569E-10 | 0 | 164716 | gtex_brain_ba24 |
| rs186969173 | 7 | 1013904 | C7orf50 | ENSG00000146540.10 | 8.002E-11 | 0 | 163992 | gtex_brain_ba24 |
| rs112587865 | 7 | 1015271 | C7orf50 | ENSG00000146540.10 | 2.561E-10 | 0 | 162625 | gtex_brain_ba24 |
| rs140499695 | 7 | 1020176 | C7orf50 | ENSG00000146540.10 | 4.579E-9 | 0 | 157720 | gtex_brain_ba24 |
| rs57492282 | 7 | 1021860 | C7orf50 | ENSG00000146540.10 | 3.971E-7 | 0 | 156036 | gtex_brain_ba24 |
| rs73259905 | 7 | 1023364 | C7orf50 | ENSG00000146540.10 | 3.073E-7 | 0 | 154532 | gtex_brain_ba24 |
| rs80133931 | 7 | 1027286 | C7orf50 | ENSG00000146540.10 | 7.524E-11 | 0 | 150610 | gtex_brain_ba24 |
| rs11438426 | 7 | 1027569 | C7orf50 | ENSG00000146540.10 | 1.485E-12 | 0 | 150327 | gtex_brain_ba24 |
| rs60892210 | 7 | 1031170 | C7orf50 | ENSG00000146540.10 | 3.44E-11 | 0 | 146726 | gtex_brain_ba24 |
| rs200978047 | 7 | 1031824 | C7orf50 | ENSG00000146540.10 | 6.693E-11 | 0 | 146072 | gtex_brain_ba24 |
| rs57858751 | 7 | 1032618 | C7orf50 | ENSG00000146540.10 | 3.951E-11 | 0 | 145278 | gtex_brain_ba24 |
| rs56109702 | 7 | 1032919 | C7orf50 | ENSG00000146540.10 | 2.041E-7 | 0 | 144977 | gtex_brain_ba24 |
| rs11977 | 7 | 1036944 | C7orf50 | ENSG00000146540.10 | 7.418E-12 | 0 | 140952 | gtex_brain_ba24 |
| rs11559183 | 7 | 1037025 | C7orf50 | ENSG00000146540.10 | 6.137E-11 | 0 | 140871 | gtex_brain_ba24 |
| rs7798636 | 7 | 1038189 | C7orf50 | ENSG00000146540.10 | 2.41E-11 | 0 | 139707 | gtex_brain_ba24 |
| rs60918895 | 7 | 1039036 | C7orf50 | ENSG00000146540.10 | 2.13E-7 | 0 | 138860 | gtex_brain_ba24 |
| rs112072378 | 7 | 1039400 | C7orf50 | ENSG00000146540.10 | 4.185E-9 | 0 | 138496 | gtex_brain_ba24 |
| rs73264010 | 7 | 1040477 | C7orf50 | ENSG00000146540.10 | 2.129E-7 | 0 | 137419 | gtex_brain_ba24 |
| rs146822086 | 7 | 1041940 | C7orf50 | ENSG00000146540.10 | 4.234E-9 | 0 | 135956 | gtex_brain_ba24 |
| rs111211004 | 7 | 1042579 | C7orf50 | ENSG00000146540.10 | 1.651E-9 | 0 | 135317 | gtex_brain_ba24 |
| rs112369387 | 7 | 1044860 | C7orf50 | ENSG00000146540.10 | 2.467E-9 | 0 | 133036 | gtex_brain_ba24 |
| rs73264028 | 7 | 1045455 | C7orf50 | ENSG00000146540.10 | 2.049E-7 | 0 | 132441 | gtex_brain_ba24 |
| rs200085503 | 7 | 1045631 | C7orf50 | ENSG00000146540.10 | 9.876E-9 | 0 | 132265 | gtex_brain_ba24 |
| rs73264032 | 7 | 1045749 | C7orf50 | ENSG00000146540.10 | 2.679E-8 | 0 | 132147 | gtex_brain_ba24 |
| rs75818004 | 7 | 1047572 | C7orf50 | ENSG00000146540.10 | 4.301E-9 | 0 | 130324 | gtex_brain_ba24 |
| rs56107018 | 7 | 1048112 | C7orf50 | ENSG00000146540.10 | 2.467E-9 | 0 | 129784 | gtex_brain_ba24 |
| rs56097140 | 7 | 1048183 | C7orf50 | ENSG00000146540.10 | 2.467E-9 | 0 | 129713 | gtex_brain_ba24 |
| rs79396168 | 7 | 1048928 | C7orf50 | ENSG00000146540.10 | 4.294E-9 | 0 | 128968 | gtex_brain_ba24 |
| rs75083636 | 7 | 1050800 | C7orf50 | ENSG00000146540.10 | 4.287E-9 | 0 | 127096 | gtex_brain_ba24 |
| rs79396675 | 7 | 1055047 | C7orf50 | ENSG00000146540.10 | 4.268E-9 | 0 | 122849 | gtex_brain_ba24 |
| rs73265918 | 7 | 1056809 | C7orf50 | ENSG00000146540.10 | 8.365E-8 | 0 | 121087 | gtex_brain_ba24 |
| rs74791540 | 7 | 1061913 | C7orf50 | ENSG00000146540.10 | 4.287E-9 | 0 | 115983 | gtex_brain_ba24 |
| rs11768895 | 7 | 1063691 | C7orf50 | ENSG00000146540.10 | 1.102E-6 | 0 | 114205 | gtex_brain_ba24 |
| rs11764817 | 7 | 1064610 | C7orf50 | ENSG00000146540.10 | 1.1E-6 | 0 | 113286 | gtex_brain_ba24 |
| rs112554101 | 7 | 1065535 | C7orf50 | ENSG00000146540.10 | 1.41E-6 | 0 | 112361 | gtex_brain_ba24 |
| rs143361185 | 7 | 1067762 | C7orf50 | ENSG00000146540.10 | 1.208E-6 | 0 | 110134 | gtex_brain_ba24 |
| rs111823776 | 7 | 1071032 | C7orf50 | ENSG00000146540.10 | 6.176E-7 | 0 | 106864 | gtex_brain_ba24 |
| rs181054388 | 7 | 1080878 | C7orf50 | ENSG00000146540.10 | 3.07E-7 | 0 | 97018 | gtex_brain_ba24 |
| rs111645884 | 7 | 1005142 | C7orf50 | ENSG00000146540.10 | 1.014E-6 | 0.01 | 172754 | gtex_brain_putamen_basal |
| rs6462277 | 7 | 1011968 | C7orf50 | ENSG00000146540.10 | 2.61E-6 | 0.01 | 165928 | gtex_brain_putamen_basal |
| rs56242247 | 7 | 1012948 | C7orf50 | ENSG00000146540.10 | 2.61E-6 | 0.01 | 164948 | gtex_brain_putamen_basal |
| rs55729719 | 7 | 1013180 | C7orf50 | ENSG00000146540.10 | 2.613E-6 | 0.01 | 164716 | gtex_brain_putamen_basal |
| rs186969173 | 7 | 1013904 | C7orf50 | ENSG00000146540.10 | 1.528E-6 | 0.01 | 163992 | gtex_brain_putamen_basal |
| rs112587865 | 7 | 1015271 | C7orf50 | ENSG00000146540.10 | 1.58E-6 | 0.01 | 162625 | gtex_brain_putamen_basal |
| rs80133931 | 7 | 1027286 | C7orf50 | ENSG00000146540.10 | 7.229E-7 | 0.01 | 150610 | gtex_brain_putamen_basal |
| rs60892210 | 7 | 1031170 | C7orf50 | ENSG00000146540.10 | 4.348E-7 | 0.01 | 146726 | gtex_brain_putamen_basal |
| rs200978047 | 7 | 1031824 | C7orf50 | ENSG00000146540.10 | 7.725E-7 | 0.01 | 146072 | gtex_brain_putamen_basal |
| rs11977 | 7 | 1036944 | C7orf50 | ENSG00000146540.10 | 1.85E-6 | 0.01 | 140952 | gtex_brain_putamen_basal |
| rs7798636 | 7 | 1038189 | C7orf50 | ENSG00000146540.10 | 1.806E-6 | 0.01 | 139707 | gtex_brain_putamen_basal |
| rs112583257 | 7 | 1091001 | C7orf50 | ENSG00000146540.10 | 1.089E-6 | 0.01 | 86895 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
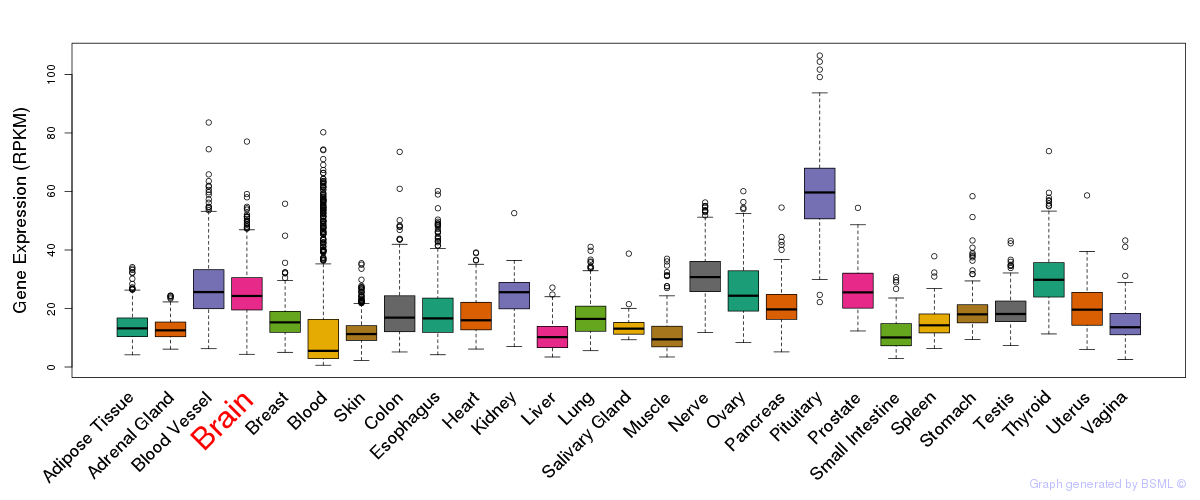
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| TONG INTERACT WITH PTTG1 | 57 | 32 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS UP | 292 | 168 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 7P22 AMPLICON | 38 | 27 | All SZGR 2.0 genes in this pathway |
| ZHU CMV 24 HR DN | 91 | 64 | All SZGR 2.0 genes in this pathway |
| ZHU CMV ALL DN | 128 | 93 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR | 149 | 84 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| BRUECKNER TARGETS OF MIRLET7A3 UP | 111 | 69 | All SZGR 2.0 genes in this pathway |
| NIELSEN GIST | 98 | 66 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |