Gene Page: MCM8
Summary ?
| GeneID | 84515 |
| Symbol | MCM8 |
| Synonyms | C20orf154|POF10|dJ967N21.5 |
| Description | minichromosome maintenance 8 homologous recombination repair factor |
| Reference | MIM:608187|HGNC:HGNC:16147|Ensembl:ENSG00000125885|HPRD:12186|Vega:OTTHUMG00000031822 |
| Gene type | protein-coding |
| Map location | 20p12.3 |
| Pascal p-value | 0.596 |
| Sherlock p-value | 0.422 |
| Fetal beta | 1.207 |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| CV:PheWAS | Phenome-wide association studies (PheWAS) | 157 SNPs associated with schizophrenia | 1 |
Section I. Genetics and epigenetics annotation
 CV:PheWAS
CV:PheWAS
| SNP ID | Chromosome | Position | Allele | P | Function | Gene | Up/Down Distance |
|---|---|---|---|---|---|---|---|
| rs16991615 | 20 | 5948227 | null | 1.883 | MCM8 | null |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs397732967 | 20 | 5932104 | MCM8 | ENSG00000125885.9 | 4.11E-7 | 0.01 | 806 | gtex_brain_ba24 |
| rs236118 | 20 | 5938207 | MCM8 | ENSG00000125885.9 | 1.435E-8 | 0.01 | 6909 | gtex_brain_ba24 |
| rs236119 | 20 | 5938654 | MCM8 | ENSG00000125885.9 | 1.068E-7 | 0.01 | 7356 | gtex_brain_ba24 |
| rs2326680 | 20 | 5942955 | MCM8 | ENSG00000125885.9 | 2.826E-7 | 0.01 | 11657 | gtex_brain_ba24 |
| rs409035 | 20 | 5943649 | MCM8 | ENSG00000125885.9 | 2.826E-7 | 0.01 | 12351 | gtex_brain_ba24 |
| rs454422 | 20 | 5943693 | MCM8 | ENSG00000125885.9 | 2.826E-7 | 0.01 | 12395 | gtex_brain_ba24 |
| rs236104 | 20 | 5958960 | MCM8 | ENSG00000125885.9 | 2.826E-7 | 0.01 | 27662 | gtex_brain_ba24 |
| rs236106 | 20 | 5960691 | MCM8 | ENSG00000125885.9 | 2.826E-7 | 0.01 | 29393 | gtex_brain_ba24 |
| rs13041190 | 20 | 5961458 | MCM8 | ENSG00000125885.9 | 1.435E-8 | 0.01 | 30160 | gtex_brain_ba24 |
| rs180477 | 20 | 5967744 | MCM8 | ENSG00000125885.9 | 2.822E-7 | 0.01 | 36446 | gtex_brain_ba24 |
| rs6053815 | 20 | 5970438 | MCM8 | ENSG00000125885.9 | 2.82E-7 | 0.01 | 39140 | gtex_brain_ba24 |
| rs6053816 | 20 | 5970750 | MCM8 | ENSG00000125885.9 | 2.819E-7 | 0.01 | 39452 | gtex_brain_ba24 |
| rs236121 | 20 | 5972594 | MCM8 | ENSG00000125885.9 | 2.818E-7 | 0.01 | 41296 | gtex_brain_ba24 |
| rs35653279 | 20 | 5974704 | MCM8 | ENSG00000125885.9 | 6.486E-7 | 0.01 | 43406 | gtex_brain_ba24 |
| rs236126 | 20 | 5978141 | MCM8 | ENSG00000125885.9 | 1.419E-8 | 0.01 | 46843 | gtex_brain_ba24 |
| rs397689224 | 20 | 5979861 | MCM8 | ENSG00000125885.9 | 5.679E-7 | 0.01 | 48563 | gtex_brain_ba24 |
| rs4813785 | 20 | 5983436 | MCM8 | ENSG00000125885.9 | 3.965E-8 | 0.01 | 52138 | gtex_brain_ba24 |
| rs6053797 | 20 | 5916096 | MCM8 | ENSG00000125885.9 | 6.144E-7 | 0 | -15202 | gtex_brain_putamen_basal |
| rs397732967 | 20 | 5932104 | MCM8 | ENSG00000125885.9 | 9.509E-10 | 0 | 806 | gtex_brain_putamen_basal |
| rs236110 | 20 | 5933108 | MCM8 | ENSG00000125885.9 | 1.022E-6 | 0 | 1810 | gtex_brain_putamen_basal |
| rs236112 | 20 | 5934319 | MCM8 | ENSG00000125885.9 | 1.022E-6 | 0 | 3021 | gtex_brain_putamen_basal |
| rs236115 | 20 | 5935449 | MCM8 | ENSG00000125885.9 | 1.022E-6 | 0 | 4151 | gtex_brain_putamen_basal |
| rs236118 | 20 | 5938207 | MCM8 | ENSG00000125885.9 | 2.304E-9 | 0 | 6909 | gtex_brain_putamen_basal |
| rs200713657 | 20 | 5938653 | MCM8 | ENSG00000125885.9 | 2.613E-9 | 0 | 7355 | gtex_brain_putamen_basal |
| rs236119 | 20 | 5938654 | MCM8 | ENSG00000125885.9 | 9.147E-12 | 0 | 7356 | gtex_brain_putamen_basal |
| rs71334368 | 20 | 5939638 | MCM8 | ENSG00000125885.9 | 6.46E-7 | 0 | 8340 | gtex_brain_putamen_basal |
| rs11483684 | 20 | 5939644 | MCM8 | ENSG00000125885.9 | 2.121E-6 | 0 | 8346 | gtex_brain_putamen_basal |
| rs236120 | 20 | 5939649 | MCM8 | ENSG00000125885.9 | 1.025E-6 | 0 | 8351 | gtex_brain_putamen_basal |
| rs379418 | 20 | 5942207 | MCM8 | ENSG00000125885.9 | 1.022E-6 | 0 | 10909 | gtex_brain_putamen_basal |
| rs2326680 | 20 | 5942955 | MCM8 | ENSG00000125885.9 | 1.153E-9 | 0 | 11657 | gtex_brain_putamen_basal |
| rs409035 | 20 | 5943649 | MCM8 | ENSG00000125885.9 | 1.153E-9 | 0 | 12351 | gtex_brain_putamen_basal |
| rs454422 | 20 | 5943693 | MCM8 | ENSG00000125885.9 | 1.153E-9 | 0 | 12395 | gtex_brain_putamen_basal |
| rs12106220 | 20 | 5944255 | MCM8 | ENSG00000125885.9 | 1.022E-6 | 0 | 12957 | gtex_brain_putamen_basal |
| rs11468712 | 20 | 5948421 | MCM8 | ENSG00000125885.9 | 8.927E-7 | 0 | 17123 | gtex_brain_putamen_basal |
| rs236102 | 20 | 5951966 | MCM8 | ENSG00000125885.9 | 1.022E-6 | 0 | 20668 | gtex_brain_putamen_basal |
| rs12624735 | 20 | 5956166 | MCM8 | ENSG00000125885.9 | 1.638E-6 | 0 | 24868 | gtex_brain_putamen_basal |
| rs236104 | 20 | 5958960 | MCM8 | ENSG00000125885.9 | 1.153E-9 | 0 | 27662 | gtex_brain_putamen_basal |
| rs236105 | 20 | 5959832 | MCM8 | ENSG00000125885.9 | 1.022E-6 | 0 | 28534 | gtex_brain_putamen_basal |
| rs236106 | 20 | 5960691 | MCM8 | ENSG00000125885.9 | 1.153E-9 | 0 | 29393 | gtex_brain_putamen_basal |
| rs13041190 | 20 | 5961458 | MCM8 | ENSG00000125885.9 | 2.304E-9 | 0 | 30160 | gtex_brain_putamen_basal |
| rs236107 | 20 | 5965731 | MCM8 | ENSG00000125885.9 | 1.022E-6 | 0 | 34433 | gtex_brain_putamen_basal |
| rs180477 | 20 | 5967744 | MCM8 | ENSG00000125885.9 | 1.143E-9 | 0 | 36446 | gtex_brain_putamen_basal |
| rs236108 | 20 | 5968559 | MCM8 | ENSG00000125885.9 | 1.022E-6 | 0 | 37261 | gtex_brain_putamen_basal |
| rs6053815 | 20 | 5970438 | MCM8 | ENSG00000125885.9 | 1.135E-9 | 0 | 39140 | gtex_brain_putamen_basal |
| rs6053816 | 20 | 5970750 | MCM8 | ENSG00000125885.9 | 1.134E-9 | 0 | 39452 | gtex_brain_putamen_basal |
| rs236121 | 20 | 5972594 | MCM8 | ENSG00000125885.9 | 1.132E-9 | 0 | 41296 | gtex_brain_putamen_basal |
| rs236122 | 20 | 5973528 | MCM8 | ENSG00000125885.9 | 6.604E-7 | 0 | 42230 | gtex_brain_putamen_basal |
| rs35653279 | 20 | 5974704 | MCM8 | ENSG00000125885.9 | 1.729E-7 | 0 | 43406 | gtex_brain_putamen_basal |
| rs236124 | 20 | 5975979 | MCM8 | ENSG00000125885.9 | 7.417E-7 | 0 | 44681 | gtex_brain_putamen_basal |
| rs236125 | 20 | 5976974 | MCM8 | ENSG00000125885.9 | 6.303E-7 | 0 | 45676 | gtex_brain_putamen_basal |
| rs236126 | 20 | 5978141 | MCM8 | ENSG00000125885.9 | 4.46E-9 | 0 | 46843 | gtex_brain_putamen_basal |
| rs397689224 | 20 | 5979861 | MCM8 | ENSG00000125885.9 | 3.302E-9 | 0 | 48563 | gtex_brain_putamen_basal |
| rs6053817 | 20 | 5980698 | MCM8 | ENSG00000125885.9 | 2.509E-7 | 0 | 49400 | gtex_brain_putamen_basal |
| rs384578 | 20 | 5981476 | MCM8 | ENSG00000125885.9 | 4.335E-7 | 0 | 50178 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
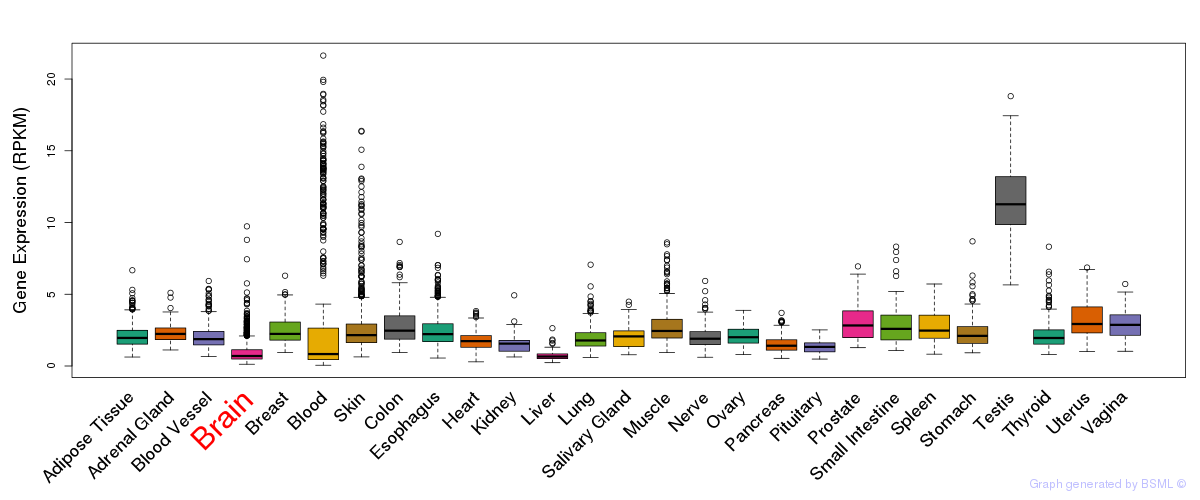
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME ACTIVATION OF THE PRE REPLICATIVE COMPLEX | 31 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE | 421 | 253 | All SZGR 2.0 genes in this pathway |
| REACTOME ORC1 REMOVAL FROM CHROMATIN | 67 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | 10 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE MITOTIC | 325 | 185 | All SZGR 2.0 genes in this pathway |
| REACTOME CELL CYCLE CHECKPOINTS | 124 | 70 | All SZGR 2.0 genes in this pathway |
| REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | 11 | 5 | All SZGR 2.0 genes in this pathway |
| REACTOME M G1 TRANSITION | 81 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME G1 S TRANSITION | 112 | 63 | All SZGR 2.0 genes in this pathway |
| REACTOME CDT1 ASSOCIATION WITH THE CDC6 ORC ORIGIN COMPLEX | 56 | 31 | All SZGR 2.0 genes in this pathway |
| REACTOME SYNTHESIS OF DNA | 92 | 53 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC G1 G1 S PHASES | 137 | 79 | All SZGR 2.0 genes in this pathway |
| REACTOME MITOTIC M M G1 PHASES | 172 | 98 | All SZGR 2.0 genes in this pathway |
| REACTOME ASSEMBLY OF THE PRE REPLICATIVE COMPLEX | 65 | 36 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA REPLICATION | 192 | 110 | All SZGR 2.0 genes in this pathway |
| REACTOME E2F MEDIATED REGULATION OF DNA REPLICATION | 35 | 17 | All SZGR 2.0 genes in this pathway |
| REACTOME ACTIVATION OF ATR IN RESPONSE TO REPLICATION STRESS | 38 | 18 | All SZGR 2.0 genes in this pathway |
| REACTOME UNWINDING OF DNA | 11 | 6 | All SZGR 2.0 genes in this pathway |
| REACTOME G2 M CHECKPOINTS | 45 | 23 | All SZGR 2.0 genes in this pathway |
| REACTOME S PHASE | 109 | 66 | All SZGR 2.0 genes in this pathway |
| REACTOME DNA STRAND ELONGATION | 30 | 18 | All SZGR 2.0 genes in this pathway |
| PYEON HPV POSITIVE TUMORS UP | 98 | 47 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA UP | 294 | 178 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED UP | 633 | 376 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 1 DN | 378 | 231 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS DN | 1024 | 594 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| KAUFFMANN DNA REPLICATION GENES | 147 | 87 | All SZGR 2.0 genes in this pathway |
| VERNELL RETINOBLASTOMA PATHWAY UP | 70 | 47 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS STEM CELL AND PROGENITOR | 681 | 420 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| TOYOTA TARGETS OF MIR34B AND MIR34C | 463 | 262 | All SZGR 2.0 genes in this pathway |
| CHANG CYCLING GENES | 148 | 83 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE S | 162 | 86 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 DN | 374 | 217 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP | 229 | 149 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 24HR UP | 324 | 193 | All SZGR 2.0 genes in this pathway |
| THILLAINADESAN ZNF217 TARGETS UP | 44 | 22 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 24HR | 128 | 73 | All SZGR 2.0 genes in this pathway |