Gene Page: PPM1D
Summary ?
| GeneID | 8493 |
| Symbol | PPM1D |
| Synonyms | PP2C-DELTA|WIP1 |
| Description | protein phosphatase, Mg2+/Mn2+ dependent 1D |
| Reference | MIM:605100|HGNC:HGNC:9277|Ensembl:ENSG00000170836|HPRD:05482|Vega:OTTHUMG00000180052 |
| Gene type | protein-coding |
| Map location | 17q23.2 |
| Pascal p-value | 0.043 |
| Fetal beta | 1.226 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg18420298 | 17 | 58677512 | PPM1D | 4.983E-4 | -0.288 | 0.047 | DMG:Wockner_2014 |
| cg10673984 | 17 | 58677844 | PPM1D | -0.026 | 0.26 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
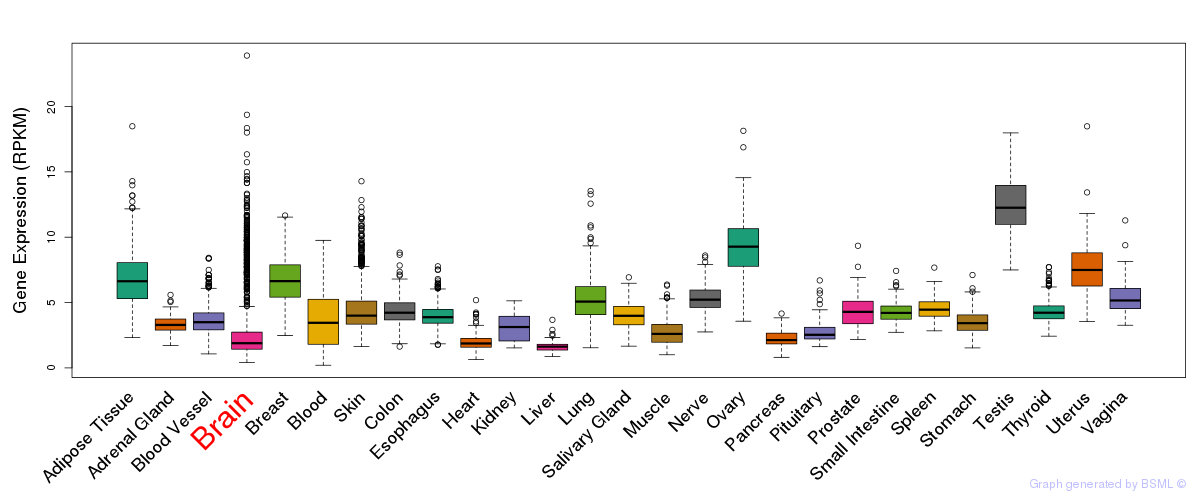
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CUL3 | 0.92 | 0.91 |
| GPBP1 | 0.91 | 0.90 |
| YME1L1 | 0.90 | 0.89 |
| SRP54 | 0.90 | 0.90 |
| CNOT7 | 0.90 | 0.90 |
| MSH2 | 0.89 | 0.88 |
| CUL2 | 0.89 | 0.89 |
| DNTTIP2 | 0.89 | 0.89 |
| DDX10 | 0.89 | 0.88 |
| DENR | 0.89 | 0.88 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.75 | -0.76 |
| AF347015.33 | -0.73 | -0.74 |
| AF347015.8 | -0.73 | -0.75 |
| AF347015.31 | -0.72 | -0.75 |
| AF347015.2 | -0.72 | -0.74 |
| MT-CYB | -0.71 | -0.73 |
| FXYD1 | -0.71 | -0.75 |
| AF347015.15 | -0.71 | -0.75 |
| AF347015.21 | -0.70 | -0.69 |
| AF347015.27 | -0.70 | -0.72 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG P53 SIGNALING PATHWAY | 69 | 45 | All SZGR 2.0 genes in this pathway |
| PID P53 REGULATION PATHWAY | 59 | 50 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| UDAYAKUMAR MED1 TARGETS DN | 240 | 171 | All SZGR 2.0 genes in this pathway |
| KIM WT1 TARGETS DN | 459 | 276 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 3 UP | 329 | 196 | All SZGR 2.0 genes in this pathway |
| KERLEY RESPONSE TO CISPLATIN UP | 44 | 30 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER UP | 181 | 108 | All SZGR 2.0 genes in this pathway |
| AMUNDSON DNA DAMAGE RESPONSE TP53 | 16 | 9 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| TSAI RESPONSE TO IONIZING RADIATION | 149 | 101 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| INGRAM SHH TARGETS DN | 64 | 41 | All SZGR 2.0 genes in this pathway |
| RICKMAN METASTASIS UP | 344 | 180 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 14HR UP | 156 | 101 | All SZGR 2.0 genes in this pathway |
| VERHAAK GLIOBLASTOMA PRONEURAL | 177 | 132 | All SZGR 2.0 genes in this pathway |
| HIRSCH CELLULAR TRANSFORMATION SIGNATURE UP | 242 | 159 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE MIDDLE | 98 | 56 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS DN | 882 | 538 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |
| MIZUSHIMA AUTOPHAGOSOME FORMATION | 19 | 15 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 2HR UP | 53 | 33 | All SZGR 2.0 genes in this pathway |
| SMIRNOV RESPONSE TO IR 6HR UP | 166 | 97 | All SZGR 2.0 genes in this pathway |
| WARTERS RESPONSE TO IR SKIN | 83 | 44 | All SZGR 2.0 genes in this pathway |
| WARTERS IR RESPONSE 5GY | 47 | 23 | All SZGR 2.0 genes in this pathway |
| ZWANG DOWN BY 2ND EGF PULSE | 293 | 119 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 5 | 18 | 11 | All SZGR 2.0 genes in this pathway |