Gene Page: IGSF21
Summary ?
| GeneID | 84966 |
| Symbol | IGSF21 |
| Synonyms | - |
| Description | immunoglobin superfamily member 21 |
| Reference | HGNC:HGNC:28246|Ensembl:ENSG00000117154|HPRD:11309|Vega:OTTHUMG00000002432 |
| Gene type | protein-coding |
| Map location | 1p36.13 |
| Pascal p-value | 4.598E-4 |
| Sherlock p-value | 0.963 |
| Fetal beta | -0.471 |
| DMG | 2 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Frontal Cortex BA9 Myers' cis & trans |
| Support | G2Cdb.humanPSD G2Cdb.humanPSP |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg13921220 | 1 | 18610522 | IGSF21 | 1.956E-4 | 0.484 | 0.035 | DMG:Wockner_2014 |
| cg17211582 | 1 | 18435858 | IGSF21 | 9.56E-9 | -0.024 | 4.24E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1578978 | chr8 | 118196802 | IGSF21 | 84966 | 0.04 | trans |
Section II. Transcriptome annotation
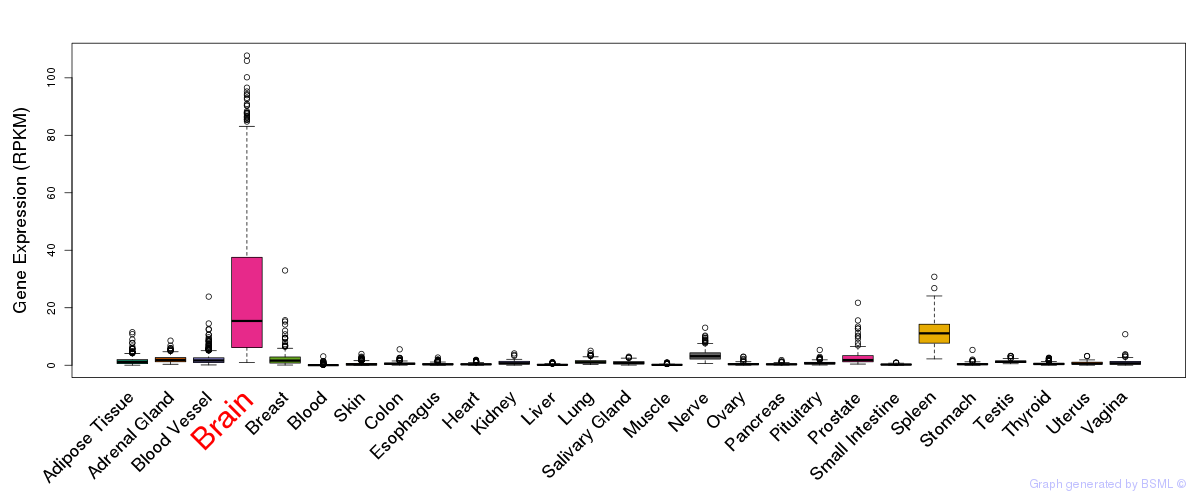
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ALDH2 | ALDH-E2 | ALDHI | ALDM | MGC1806 | aldehyde dehydrogenase 2 family (mitochondrial) | Two-hybrid | BioGRID | 16169070 |
| ANXA3 | ANX3 | annexin A3 | Two-hybrid | BioGRID | 16169070 |
| ATF3 | - | activating transcription factor 3 | Two-hybrid | BioGRID | 16169070 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | Two-hybrid | BioGRID | 16169070 |
| BTBD2 | - | BTB (POZ) domain containing 2 | Two-hybrid | BioGRID | 16169070 |
| C5orf13 | D4S114 | P311 | PRO1873 | PTZ17 | chromosome 5 open reading frame 13 | Two-hybrid | BioGRID | 16169070 |
| CRCT1 | C1orf42 | NICE-1 | cysteine-rich C-terminal 1 | Two-hybrid | BioGRID | 16169070 |
| DGKD | DGKdelta | FLJ26930 | KIAA0145 | dgkd-2 | diacylglycerol kinase, delta 130kDa | Two-hybrid | BioGRID | 16169070 |
| ERH | DROER | FLJ27340 | enhancer of rudimentary homolog (Drosophila) | Two-hybrid | BioGRID | 16169070 |
| ETHE1 | HSCO | YF13H12 | ethylmalonic encephalopathy 1 | Two-hybrid | BioGRID | 16169070 |
| GIPC2 | FLJ20075 | SEMCAP-2 | SEMCAP2 | GIPC PDZ domain containing family, member 2 | Two-hybrid | BioGRID | 16169070 |
| GSTM4 | GSTM4-4 | GTM4 | MGC131945 | MGC9247 | glutathione S-transferase mu 4 | Two-hybrid | BioGRID | 16169070 |
| HSPB1 | CMT2F | DKFZp586P1322 | HMN2B | HS.76067 | HSP27 | HSP28 | Hsp25 | SRP27 | heat shock 27kDa protein 1 | Two-hybrid | BioGRID | 16169070 |
| MAD2L1BP | CMT2 | KIAA0110 | MGC11282 | RP1-261G23.6 | MAD2L1 binding protein | Two-hybrid | BioGRID | 16169070 |
| NACA | HSD48 | MGC117224 | NACA1 | nascent polypeptide-associated complex alpha subunit | Two-hybrid | BioGRID | 16169070 |
| PAEP | GD | GdA | GdF | GdS | MGC138509 | MGC142288 | PAEG | PEP | PP14 | progestagen-associated endometrial protein | Two-hybrid | BioGRID | 16169070 |
| PAFAH1B3 | - | platelet-activating factor acetylhydrolase, isoform Ib, gamma subunit 29kDa | Two-hybrid | BioGRID | 16169070 |
| PCDHA4 | CNR1 | CNRN1 | CRNR1 | MGC138307 | MGC142169 | PCDH-ALPHA4 | protocadherin alpha 4 | Two-hybrid | BioGRID | 16169070 |
| PRMT1 | ANM1 | HCP1 | HRMT1L2 | IR1B4 | protein arginine methyltransferase 1 | Two-hybrid | BioGRID | 16169070 |
| PSMD11 | MGC3844 | Rpn6 | S9 | p44.5 | proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 | Two-hybrid | BioGRID | 16169070 |
| RPP14 | FLJ31508 | P14 | ribonuclease P/MRP 14kDa subunit | Two-hybrid | BioGRID | 16169070 |
| S100A8 | 60B8AG | CAGA | CFAG | CGLA | CP-10 | L1Ag | MA387 | MIF | MRP8 | NIF | P8 | S100 calcium binding protein A8 | Two-hybrid | BioGRID | 16169070 |
| SEPHS1 | MGC4980 | SELD | SPS | SPS1 | selenophosphate synthetase 1 | Two-hybrid | BioGRID | 16169070 |
| SERPINB9 | CAP-3 | CAP3 | PI9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | Two-hybrid | BioGRID | 16169070 |
| SULT1E1 | EST | EST-1 | MGC34459 | STE | sulfotransferase family 1E, estrogen-preferring, member 1 | Two-hybrid | BioGRID | 16169070 |
| TRBV2 | TCRBV22S1A2N1T | TCRBV2S1 | T cell receptor beta variable 2 | Two-hybrid | BioGRID | 16169070 |
| UBE2V2 | DDVIT1 | DDVit-1 | EDAF-1 | EDPF-1 | EDPF1 | MMS2 | UEV-2 | UEV2 | ubiquitin-conjugating enzyme E2 variant 2 | Two-hybrid | BioGRID | 16169070 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| BENPORATH SUZ12 TARGETS | 1038 | 678 | All SZGR 2.0 genes in this pathway |
| BENPORATH EED TARGETS | 1062 | 725 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES WITH H3K27ME3 | 1118 | 744 | All SZGR 2.0 genes in this pathway |
| BENPORATH PRC2 TARGETS | 652 | 441 | All SZGR 2.0 genes in this pathway |
| WHITEHURST PACLITAXEL SENSITIVITY | 39 | 19 | All SZGR 2.0 genes in this pathway |
| HANN RESISTANCE TO BCL2 INHIBITOR DN | 48 | 31 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 AND H3K27ME3 | 349 | 234 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN NPC HCP WITH H3K27ME3 | 341 | 243 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MEF HCP WITH H3K27ME3 | 590 | 403 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |