Gene Page: RANBP3
Summary ?
| GeneID | 8498 |
| Symbol | RANBP3 |
| Synonyms | - |
| Description | RAN binding protein 3 |
| Reference | MIM:603327|HGNC:HGNC:9850|Ensembl:ENSG00000031823|HPRD:04508|Vega:OTTHUMG00000180636 |
| Gene type | protein-coding |
| Map location | 19p13.3 |
| Pascal p-value | 0.461 |
| Fetal beta | 0.548 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14737613 | 19 | 5978790 | RANBP3 | 4.26E-9 | -0.029 | 2.55E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
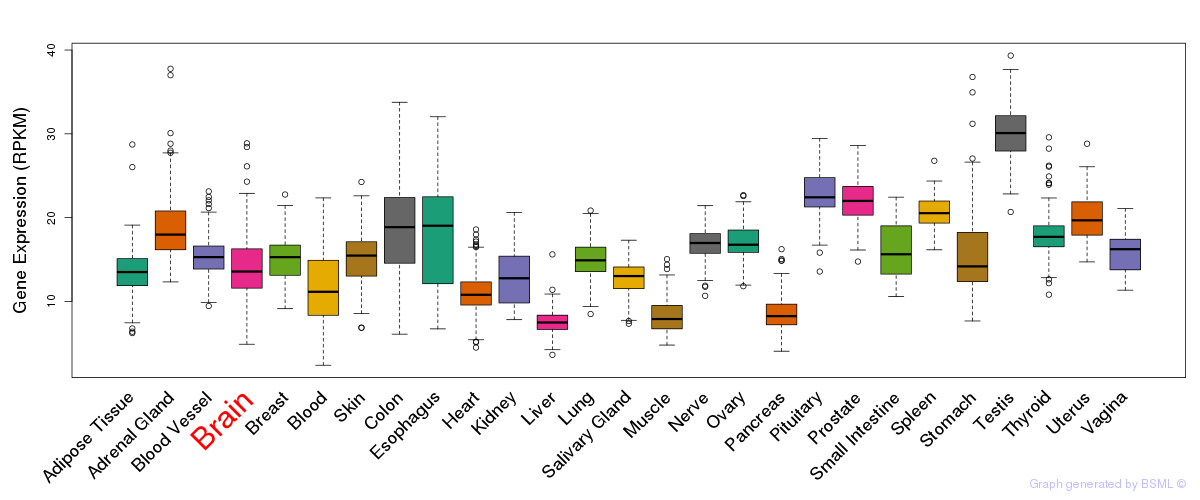
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MDH1 | 0.77 | 0.71 |
| EPHX4 | 0.74 | 0.69 |
| CHN1 | 0.74 | 0.75 |
| NRSN1 | 0.73 | 0.65 |
| LYSMD2 | 0.72 | 0.68 |
| GSTO1 | 0.72 | 0.76 |
| ACYP2 | 0.71 | 0.72 |
| ABCC12 | 0.71 | 0.67 |
| FBXO9 | 0.70 | 0.63 |
| VIP | 0.70 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| FADS2 | -0.61 | -0.65 |
| SEMA4B | -0.57 | -0.63 |
| KIAA1949 | -0.56 | -0.61 |
| PKN1 | -0.56 | -0.63 |
| TUBB2B | -0.56 | -0.66 |
| SH3BP2 | -0.55 | -0.69 |
| KCTD11 | -0.55 | -0.66 |
| GLIS2 | -0.55 | -0.64 |
| FAM129B | -0.54 | -0.63 |
| JUP | -0.54 | -0.58 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID WNT CANONICAL PATHWAY | 20 | 18 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN DN | 241 | 146 | All SZGR 2.0 genes in this pathway |
| HORIUCHI WTAP TARGETS UP | 306 | 188 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| EBAUER TARGETS OF PAX3 FOXO1 FUSION UP | 207 | 128 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 60 MCF10A | 39 | 24 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS DN | 61 | 47 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A5 | 70 | 32 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 NOT SATB1 UP | 344 | 215 | All SZGR 2.0 genes in this pathway |