Gene Page: PEX3
Summary ?
| GeneID | 8504 |
| Symbol | PEX3 |
| Synonyms | PBD10A|TRG18 |
| Description | peroxisomal biogenesis factor 3 |
| Reference | MIM:603164|HGNC:HGNC:8858|HPRD:04407| |
| Gene type | protein-coding |
| Map location | 6q24.2 |
| Pascal p-value | 0.646 |
| Fetal beta | -0.121 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06866400 | 6 | 143771904 | ADAT2;PEX3 | 1.33E-5 | -0.375 | 0.014 | DMG:Wockner_2014 |
| cg21684385 | 6 | 143772210 | PEX3 | 1.07E-8 | -0.014 | 4.56E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
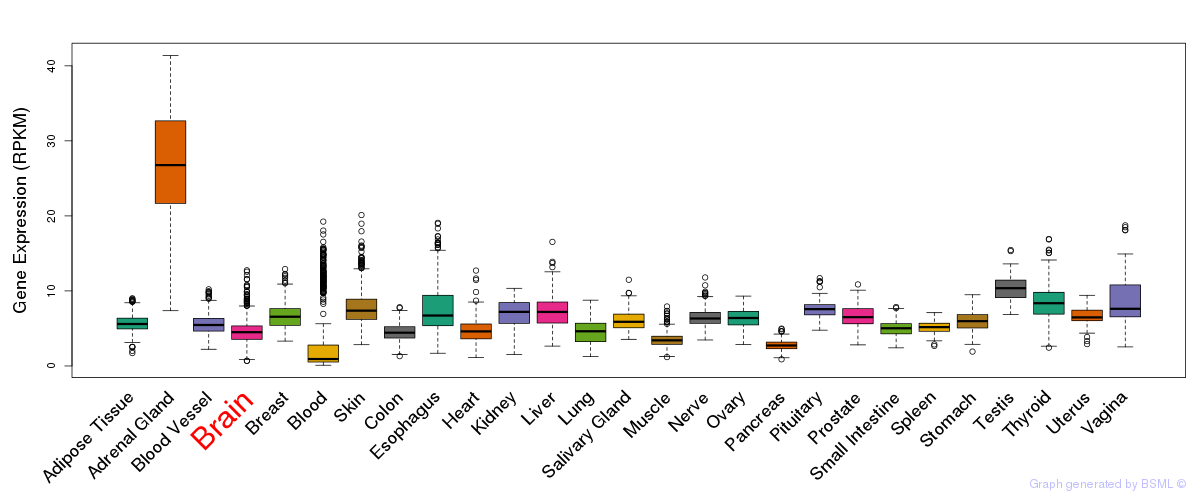
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SF1 | 0.96 | 0.96 |
| DNMT1 | 0.95 | 0.96 |
| CHERP | 0.95 | 0.97 |
| RPRD2 | 0.95 | 0.94 |
| CPSF7 | 0.94 | 0.93 |
| NUP214 | 0.94 | 0.94 |
| PFAS | 0.94 | 0.96 |
| PATL1 | 0.94 | 0.92 |
| MEN1 | 0.94 | 0.95 |
| SF3A1 | 0.94 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.78 | -0.89 |
| MT-CO2 | -0.77 | -0.90 |
| C5orf53 | -0.75 | -0.79 |
| AF347015.33 | -0.74 | -0.84 |
| S100B | -0.74 | -0.84 |
| AF347015.27 | -0.74 | -0.85 |
| FXYD1 | -0.74 | -0.84 |
| MT-CYB | -0.73 | -0.84 |
| HIGD1B | -0.72 | -0.85 |
| AF347015.8 | -0.72 | -0.86 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IPI | 10704444 |11883941 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007031 | peroxisome organization | IEA | - | |
| GO:0007031 | peroxisome organization | IMP | 10958759 |12924628 | |
| GO:0045046 | protein import into peroxisome membrane | IMP | 15007061 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005777 | peroxisome | IMP | 12924628 | |
| GO:0005778 | peroxisomal membrane | IEA | - | |
| GO:0005779 | integral to peroxisomal membrane | IDA | 9657383 |10430017 | |
| GO:0005779 | integral to peroxisomal membrane | IEA | - | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PEROXISOME | 78 | 47 | All SZGR 2.0 genes in this pathway |
| REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | 18 | 11 | All SZGR 2.0 genes in this pathway |
| REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | 34 | 21 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSMEMBRANE TRANSPORT OF SMALL MOLECULES | 413 | 270 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING DN | 58 | 35 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER APOCRINE VS LUMINAL | 326 | 213 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC TARGETS WITH EBOX | 230 | 156 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS UP | 424 | 268 | All SZGR 2.0 genes in this pathway |
| FERNANDEZ BOUND BY MYC | 182 | 116 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT REJECTED VS OK DN | 546 | 351 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| MCCABE BOUND BY HOXC6 | 469 | 239 | All SZGR 2.0 genes in this pathway |
| ACEVEDO NORMAL TISSUE ADJACENT TO LIVER TUMOR UP | 174 | 96 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE DN | 274 | 165 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER POOR SURVIVAL UP | 31 | 22 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE LATE | 1137 | 655 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND WITH H4K20ME1 MARK | 145 | 82 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FARMER BREAST CANCER CLUSTER 3 | 16 | 12 | All SZGR 2.0 genes in this pathway |