Gene Page: NKD2
Summary ?
| GeneID | 85409 |
| Symbol | NKD2 |
| Synonyms | Naked2 |
| Description | naked cuticle homolog 2 (Drosophila) |
| Reference | MIM:607852|HGNC:HGNC:17046|Ensembl:ENSG00000145506|HPRD:08483|Vega:OTTHUMG00000090348 |
| Gene type | protein-coding |
| Map location | 5p15.3 |
| Pascal p-value | 0.66 |
| Sherlock p-value | 0.059 |
| Fetal beta | -0.157 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Cortex Hypothalamus Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15165927 | 5 | 1033518 | NKD2 | 2.207E-4 | 0.418 | 0.036 | DMG:Wockner_2014 |
| cg26489368 | 5 | 1033593 | NKD2 | 3.116E-4 | 0.53 | 0.04 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1111480 | chr16 | 52521355 | NKD2 | 85409 | 0.09 | trans | ||
| rs7188610 | chr16 | 52527281 | NKD2 | 85409 | 0.12 | trans | ||
| rs56271444 | 5 | 1021789 | NKD2 | ENSG00000145506.9 | 1.607E-7 | 0.01 | 12845 | gtex_brain_ba24 |
| rs56006100 | 5 | 1022326 | NKD2 | ENSG00000145506.9 | 5.283E-7 | 0.01 | 13382 | gtex_brain_ba24 |
| rs72707203 | 5 | 1026720 | NKD2 | ENSG00000145506.9 | 4.48E-7 | 0.01 | 17776 | gtex_brain_ba24 |
| rs1864094 | 5 | 1027456 | NKD2 | ENSG00000145506.9 | 2.534E-7 | 0.01 | 18512 | gtex_brain_ba24 |
| rs10037180 | 5 | 1027473 | NKD2 | ENSG00000145506.9 | 6.334E-8 | 0.01 | 18529 | gtex_brain_ba24 |
| rs10045604 | 5 | 1028683 | NKD2 | ENSG00000145506.9 | 1.538E-7 | 0.01 | 19739 | gtex_brain_ba24 |
| rs11743637 | 5 | 1028918 | NKD2 | ENSG00000145506.9 | 2.536E-7 | 0.01 | 19974 | gtex_brain_ba24 |
| rs1560174 | 5 | 1029215 | NKD2 | ENSG00000145506.9 | 2.534E-7 | 0.01 | 20271 | gtex_brain_ba24 |
| rs397814216 | 5 | 1029243 | NKD2 | ENSG00000145506.9 | 6.085E-8 | 0.01 | 20299 | gtex_brain_ba24 |
| rs882622 | 5 | 1029712 | NKD2 | ENSG00000145506.9 | 2.364E-7 | 0.01 | 20768 | gtex_brain_ba24 |
| rs882623 | 5 | 1029851 | NKD2 | ENSG00000145506.9 | 5.54E-7 | 0.01 | 20907 | gtex_brain_ba24 |
| rs199825697 | 5 | 1030136 | NKD2 | ENSG00000145506.9 | 2.886E-7 | 0.01 | 21192 | gtex_brain_ba24 |
| rs201625977 | 5 | 1030141 | NKD2 | ENSG00000145506.9 | 2.156E-7 | 0.01 | 21197 | gtex_brain_ba24 |
| rs881334 | 5 | 1030645 | NKD2 | ENSG00000145506.9 | 2.093E-7 | 0.01 | 21701 | gtex_brain_ba24 |
| rs10061048 | 5 | 1030972 | NKD2 | ENSG00000145506.9 | 6.193E-8 | 0.01 | 22028 | gtex_brain_ba24 |
| rs4975577 | 5 | 1032032 | NKD2 | ENSG00000145506.9 | 6.195E-8 | 0.01 | 23088 | gtex_brain_ba24 |
| rs4975576 | 5 | 1034246 | NKD2 | ENSG00000145506.9 | 2.646E-7 | 0.01 | 25302 | gtex_brain_ba24 |
| rs34309815 | 5 | 1036900 | NKD2 | ENSG00000145506.9 | 4.516E-7 | 0.01 | 27956 | gtex_brain_ba24 |
| rs12517757 | 5 | 1040657 | NKD2 | ENSG00000145506.9 | 1.273E-7 | 0.01 | 31713 | gtex_brain_ba24 |
| rs11739445 | 5 | 1043397 | NKD2 | ENSG00000145506.9 | 1.273E-7 | 0.01 | 34453 | gtex_brain_ba24 |
| rs146026905 | 5 | 1043524 | NKD2 | ENSG00000145506.9 | 5.651E-7 | 0.01 | 34580 | gtex_brain_ba24 |
| rs36193315 | 5 | 1047394 | NKD2 | ENSG00000145506.9 | 1.423E-7 | 0.01 | 38450 | gtex_brain_ba24 |
| rs76996633 | 5 | 1047471 | NKD2 | ENSG00000145506.9 | 2.333E-7 | 0.01 | 38527 | gtex_brain_ba24 |
| rs150880358 | 5 | 1047655 | NKD2 | ENSG00000145506.9 | 2.123E-7 | 0.01 | 38711 | gtex_brain_ba24 |
| rs376871773 | 5 | 1047694 | NKD2 | ENSG00000145506.9 | 1.252E-7 | 0.01 | 38750 | gtex_brain_ba24 |
| rs62329235 | 5 | 1049091 | NKD2 | ENSG00000145506.9 | 1.305E-7 | 0.01 | 40147 | gtex_brain_ba24 |
| rs55739458 | 5 | 1049186 | NKD2 | ENSG00000145506.9 | 1.273E-7 | 0.01 | 40242 | gtex_brain_ba24 |
| rs55817553 | 5 | 1049351 | NKD2 | ENSG00000145506.9 | 1.268E-7 | 0.01 | 40407 | gtex_brain_ba24 |
| rs34746201 | 5 | 1049625 | NKD2 | ENSG00000145506.9 | 3.115E-7 | 0.01 | 40681 | gtex_brain_ba24 |
| rs145524812 | 5 | 1049635 | NKD2 | ENSG00000145506.9 | 5.837E-7 | 0.01 | 40691 | gtex_brain_ba24 |
| rs34642520 | 5 | 1049642 | NKD2 | ENSG00000145506.9 | 5.525E-7 | 0.01 | 40698 | gtex_brain_ba24 |
| rs13190524 | 5 | 1049694 | NKD2 | ENSG00000145506.9 | 1.286E-7 | 0.01 | 40750 | gtex_brain_ba24 |
| rs1058508 | 5 | 1051687 | NKD2 | ENSG00000145506.9 | 5.428E-7 | 0.01 | 42743 | gtex_brain_ba24 |
| rs79562755 | 5 | 1052735 | NKD2 | ENSG00000145506.9 | 1.261E-7 | 0.01 | 43791 | gtex_brain_ba24 |
| rs2241601 | 5 | 1053216 | NKD2 | ENSG00000145506.9 | 1.268E-7 | 0.01 | 44272 | gtex_brain_ba24 |
| rs2241602 | 5 | 1053341 | NKD2 | ENSG00000145506.9 | 5.409E-7 | 0.01 | 44397 | gtex_brain_ba24 |
| rs2241607 | 5 | 1057890 | NKD2 | ENSG00000145506.9 | 4.648E-7 | 0.01 | 48946 | gtex_brain_ba24 |
| rs2241608 | 5 | 1060685 | NKD2 | ENSG00000145506.9 | 4.778E-7 | 0.01 | 51741 | gtex_brain_ba24 |
| rs4438919 | 5 | 1061688 | NKD2 | ENSG00000145506.9 | 5.143E-7 | 0.01 | 52744 | gtex_brain_ba24 |
| rs3789198 | 5 | 1067483 | NKD2 | ENSG00000145506.9 | 5.089E-7 | 0.01 | 58539 | gtex_brain_ba24 |
| rs4073269 | 5 | 1087868 | NKD2 | ENSG00000145506.9 | 4.702E-7 | 0.01 | 78924 | gtex_brain_ba24 |
Section II. Transcriptome annotation
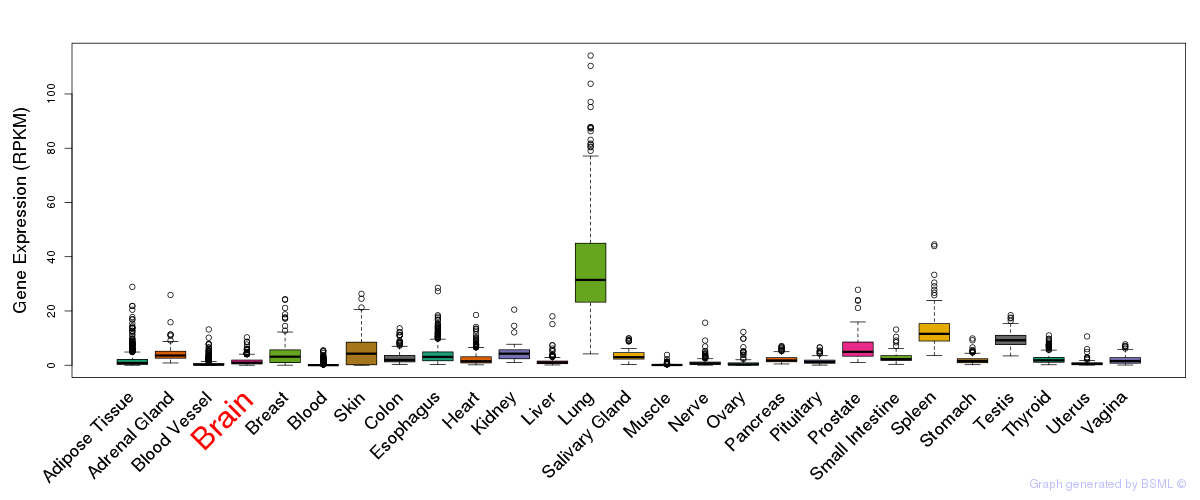
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG WNT SIGNALING PATHWAY | 151 | 112 | All SZGR 2.0 genes in this pathway |
| ST WNT BETA CATENIN PATHWAY | 34 | 28 | All SZGR 2.0 genes in this pathway |
| PID WNT CANONICAL PATHWAY | 20 | 18 | All SZGR 2.0 genes in this pathway |
| SABATES COLORECTAL ADENOMA SIZE UP | 23 | 12 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| THEODOROU MAMMARY TUMORIGENESIS | 31 | 24 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 5P15 AMPLICON | 26 | 15 | All SZGR 2.0 genes in this pathway |
| CHEN NEUROBLASTOMA COPY NUMBER GAINS | 50 | 35 | All SZGR 2.0 genes in this pathway |
| ROZANOV MMP14 TARGETS UP | 266 | 171 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 TARGETS UP | 673 | 430 | All SZGR 2.0 genes in this pathway |
| MARTINEZ TP53 TARGETS DN | 593 | 372 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RB1 AND TP53 TARGETS DN | 591 | 366 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN MCV6 HCP WITH H3K27ME3 | 435 | 318 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP | 745 | 475 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION UP | 570 | 339 | All SZGR 2.0 genes in this pathway |