Gene Page: TNFSF11
Summary ?
| GeneID | 8600 |
| Symbol | TNFSF11 |
| Synonyms | CD254|ODF|OPGL|OPTB2|RANKL|TNLG6B|TRANCE|hRANKL2|sOdf |
| Description | tumor necrosis factor superfamily member 11 |
| Reference | MIM:602642|HGNC:HGNC:11926|Ensembl:ENSG00000120659|HPRD:04031|Vega:OTTHUMG00000016807 |
| Gene type | protein-coding |
| Map location | 13q14 |
| Pascal p-value | 0.718 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0312 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24534742 | 13 | 43149281 | TNFSF11 | 2.266E-4 | -0.233 | 0.036 | DMG:Wockner_2014 |
Section II. Transcriptome annotation
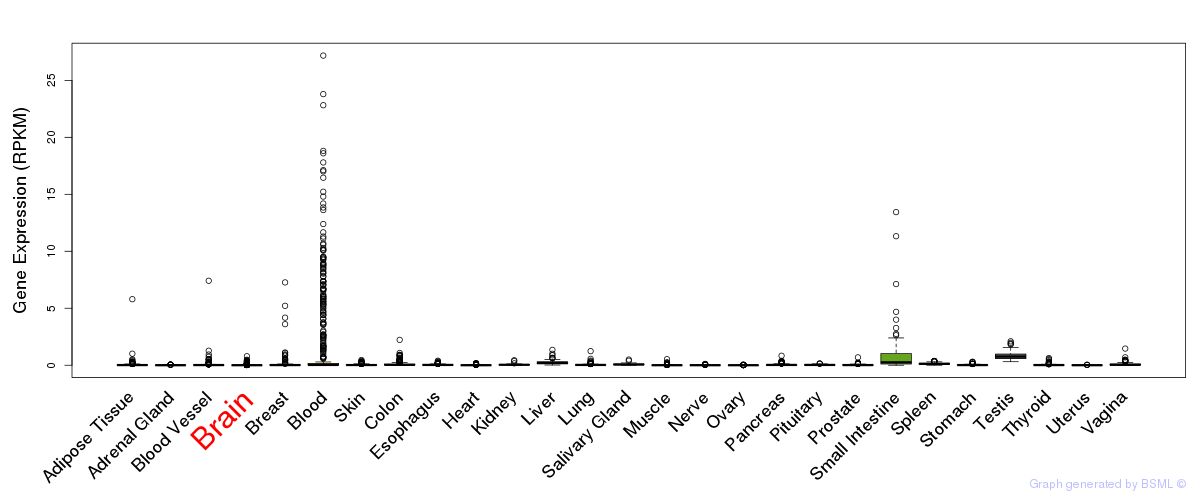
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DNAJB6 | 0.92 | 0.89 |
| SUMO1 | 0.90 | 0.83 |
| SSB | 0.89 | 0.87 |
| CBX3 | 0.88 | 0.85 |
| DENR | 0.88 | 0.84 |
| DNTTIP2 | 0.88 | 0.84 |
| FIP1L1 | 0.87 | 0.87 |
| YWHAE | 0.87 | 0.83 |
| SNX4 | 0.87 | 0.81 |
| RALA | 0.87 | 0.87 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TREX1 | -0.62 | -0.73 |
| HLA-F | -0.62 | -0.70 |
| FXYD1 | -0.61 | -0.70 |
| TSC22D4 | -0.60 | -0.70 |
| MT-CO2 | -0.60 | -0.68 |
| AC018755.7 | -0.60 | -0.66 |
| AF347015.33 | -0.59 | -0.68 |
| IFI27 | -0.59 | -0.70 |
| AF347015.9 | -0.59 | -0.72 |
| AF347015.31 | -0.58 | -0.68 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004872 | receptor activity | IEA | - | |
| GO:0005125 | cytokine activity | NAS | 9568710 | |
| GO:0005164 | tumor necrosis factor receptor binding | NAS | 9312132 |9367155 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006955 | immune response | NAS | 9312132 | |
| GO:0007275 | multicellular organismal development | IEA | - | |
| GO:0030154 | cell differentiation | IEA | - | |
| GO:0045780 | positive regulation of bone resorption | IDA | 17241109 | |
| GO:0045672 | positive regulation of osteoclast differentiation | IDA | 17241109 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005576 | extracellular region | NAS | 9568710 |10708588 | |
| GO:0005615 | extracellular space | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | NAS | 9312132 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AKT1 | AKT | MGC99656 | PKB | PKB-ALPHA | PRKBA | RAC | RAC-ALPHA | v-akt murine thymoma viral oncogene homolog 1 | - | HPRD | 10635328 |
| KHDRBS1 | FLJ34027 | Sam68 | p62 | KH domain containing, RNA binding, signal transduction associated 1 | Biochemical Activity | BioGRID | 10635328 |
| MAPK1 | ERK | ERK2 | ERT1 | MAPK2 | P42MAPK | PRKM1 | PRKM2 | p38 | p40 | p41 | p41mapk | mitogen-activated protein kinase 1 | - | HPRD | 10635328 |
| MAPK3 | ERK1 | HS44KDAP | HUMKER1A | MGC20180 | P44ERK1 | P44MAPK | PRKM3 | mitogen-activated protein kinase 3 | - | HPRD | 10635328 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | - | HPRD | 9396779 |
| NFKB1 | DKFZp686C01211 | EBP-1 | KBF1 | MGC54151 | NF-kappa-B | NFKB-p105 | NFKB-p50 | p105 | nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 | - | HPRD | 10635328 |
| NFKBIA | IKBA | MAD-3 | NFKBI | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha | - | HPRD | 10635328 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 10635328 |
| TNFRSF11A | CD265 | FEO | LOH18CR1 | ODFR | OFE | OPTB7 | OSTS | PDB2 | RANK | TRANCER | tumor necrosis factor receptor superfamily, member 11a, NFKB activator | - | HPRD | 9367155 |
| TNFRSF11B | MGC29565 | OCIF | OPG | TR1 | tumor necrosis factor receptor superfamily, member 11b | - | HPRD,BioGRID | 9520411|12221720 |
| TRAF6 | MGC:3310 | RNF85 | TNF receptor-associated factor 6 | - | HPRD | 10635328 |
| ZNF675 | FLJ36350 | TBZF | TIZ | zinc finger protein 675 | Phenotypic Suppression | BioGRID | 11751921 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG CYTOKINE CYTOKINE RECEPTOR INTERACTION | 267 | 161 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RANKL PATHWAY | 14 | 13 | All SZGR 2.0 genes in this pathway |
| PID IL6 7 PATHWAY | 47 | 40 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LOPEZ MBD TARGETS | 957 | 597 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| BROWN MYELOID CELL DEVELOPMENT DN | 129 | 86 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN | 234 | 137 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| SAFFORD T LYMPHOCYTE ANERGY | 87 | 54 | All SZGR 2.0 genes in this pathway |
| ZHENG BOUND BY FOXP3 | 491 | 310 | All SZGR 2.0 genes in this pathway |
| ZHENG FOXP3 TARGETS IN THYMUS UP | 196 | 137 | All SZGR 2.0 genes in this pathway |
| WORSCHECH TUMOR REJECTION UP | 56 | 32 | All SZGR 2.0 genes in this pathway |
| QI PLASMACYTOMA UP | 259 | 185 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME2 AND H3K27ME3 | 59 | 35 | All SZGR 2.0 genes in this pathway |
| MEISSNER NPC HCP WITH H3K4ME2 | 491 | 319 | All SZGR 2.0 genes in this pathway |
| MARSON FOXP3 TARGETS DN | 54 | 38 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 30MIN DN | 150 | 99 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| FUKUSHIMA TNFSF11 TARGETS | 16 | 14 | All SZGR 2.0 genes in this pathway |
| CARD MIR302A TARGETS | 77 | 62 | All SZGR 2.0 genes in this pathway |
| PURBEY TARGETS OF CTBP1 AND SATB1 DN | 180 | 116 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY LUMINAL MATURE UP | 116 | 65 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |
| NABA SECRETED FACTORS | 344 | 197 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME ASSOCIATED | 753 | 411 | All SZGR 2.0 genes in this pathway |
| NABA MATRISOME | 1028 | 559 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-124/506 | 801 | 807 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-17-5p/20/93.mr/106/519.d | 617 | 623 | m8 | hsa-miR-17-5p | CAAAGUGCUUACAGUGCAGGUAGU |
| hsa-miR-20abrain | UAAAGUGCUUAUAGUGCAGGUAG | ||||
| hsa-miR-106a | AAAAGUGCUUACAGUGCAGGUAGC | ||||
| hsa-miR-106bSZ | UAAAGUGCUGACAGUGCAGAU | ||||
| hsa-miR-20bSZ | CAAAGUGCUCAUAGUGCAGGUAG | ||||
| hsa-miR-519d | CAAAGUGCCUCCCUUUAGAGUGU | ||||
| miR-181 | 1049 | 1055 | m8 | hsa-miR-181abrain | AACAUUCAACGCUGUCGGUGAGU |
| hsa-miR-181bSZ | AACAUUCAUUGCUGUCGGUGGG | ||||
| hsa-miR-181cbrain | AACAUUCAACCUGUCGGUGAGU | ||||
| hsa-miR-181dbrain | AACAUUCAUUGUUGUCGGUGGGUU | ||||
| miR-194 | 398 | 404 | 1A | hsa-miR-194 | UGUAACAGCAACUCCAUGUGGA |
| miR-410 | 463 | 469 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| hsa-miR-410 | AAUAUAACACAGAUGGCCUGU | ||||
| miR-493-5p | 594 | 600 | m8 | hsa-miR-493-5p | UUGUACAUGGUAGGCUUUCAUU |
| miR-543 | 1050 | 1057 | 1A,m8 | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.