Gene Page: RUNX1T1
Summary ?
| GeneID | 862 |
| Symbol | RUNX1T1 |
| Synonyms | AML1-MTG8|AML1T1|CBFA2T1|CDR|ETO|MTG8|ZMYND2 |
| Description | runt related transcription factor 1; translocated to, 1 (cyclin D related) |
| Reference | MIM:133435|HGNC:HGNC:1535|Ensembl:ENSG00000079102|HPRD:00590|Vega:OTTHUMG00000164066 |
| Gene type | protein-coding |
| Map location | 8q22 |
| Pascal p-value | 0.698 |
| Sherlock p-value | 0.024 |
| Fetal beta | 3.241 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg00045118 | 8 | 93076177 | RUNX1T1 | 2.381E-4 | 0.438 | 0.037 | DMG:Wockner_2014 |
| cg11043251 | 8 | 93108262 | RUNX1T1 | 5.174E-4 | 0.444 | 0.047 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs17686978 | chr8 | 93725522 | RUNX1T1 | 862 | 0.13 | cis | ||
| rs16829545 | chr2 | 151977407 | RUNX1T1 | 862 | 0.12 | trans |
Section II. Transcriptome annotation
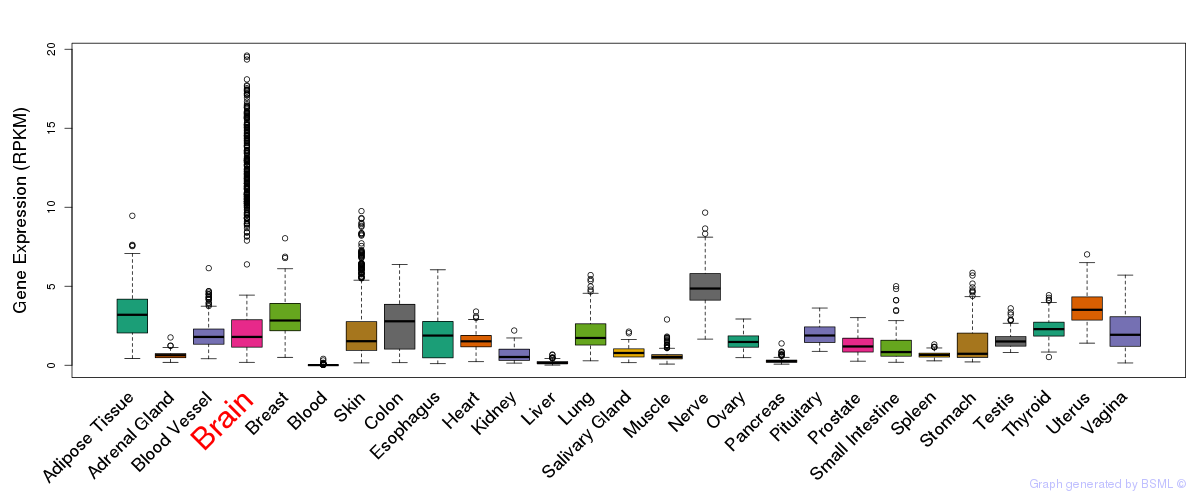
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABI3 | NESH | SSH3BP3 | ABI family, member 3 | Two-hybrid | BioGRID | 16189514 |
| C19orf57 | MGC11271 | MGC149720 | chromosome 19 open reading frame 57 | Two-hybrid | BioGRID | 16189514 |
| CBFA2T2 | DKFZp313F2116 | EHT | MTGR1 | ZMYND3 | core-binding factor, runt domain, alpha subunit 2; translocated to, 2 | ETO interacts with MTGR1. | BIND | 15333839 |
| CBFA2T2 | DKFZp313F2116 | EHT | MTGR1 | ZMYND3 | core-binding factor, runt domain, alpha subunit 2; translocated to, 2 | - | HPRD | 10675041 |
| CBFA2T2 | DKFZp313F2116 | EHT | MTGR1 | ZMYND3 | core-binding factor, runt domain, alpha subunit 2; translocated to, 2 | Affinity Capture-Western | BioGRID | 12242670 |14703694 |16189514 |
| CBFA2T3 | ETO2 | MTG16 | MTGR2 | ZMYND4 | core-binding factor, runt domain, alpha subunit 2; translocated to, 3 | - | HPRD,BioGRID | 14703694 |
| CEP170L | FAM68B | KIAA0470 | KIAA0470L | MGC26143 | centrosomal protein 170kDa-like | Two-hybrid | BioGRID | 16189514 |
| GFI1 | FLJ94509 | GFI-1 | ZNF163 | growth factor independent 1 transcription repressor | - | HPRD,BioGRID | 12874834 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | Affinity Capture-Western | BioGRID | 11533236 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | ETO interacts with HDAC1. | BIND | 15333839 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-Western | BioGRID | 11533236 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | Affinity Capture-Western | BioGRID | 11533236 |
| HDAC3 | HD3 | RPD3 | RPD3-2 | histone deacetylase 3 | ETO interacts with HDAC3. | BIND | 15333839 |
| HDAC8 | HDACL1 | RPD3 | histone deacetylase 8 | - | HPRD,BioGRID | 11533236 |
| HSP90AA1 | FLJ31884 | HSP86 | HSP89A | HSP90A | HSP90N | HSPC1 | HSPCA | HSPCAL1 | HSPCAL4 | HSPN | Hsp89 | Hsp90 | LAP2 | heat shock protein 90kDa alpha (cytosolic), class A member 1 | - | HPRD | 10076566 |
| KPNA1 | IPOA5 | NPI-1 | RCH2 | SRP1 | karyopherin alpha 1 (importin alpha 5) | - | HPRD,BioGRID | 10951564 |
| KPNB1 | IMB1 | IPOB | Impnb | MGC2155 | MGC2156 | MGC2157 | NTF97 | karyopherin (importin) beta 1 | - | HPRD,BioGRID | 10951564 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | - | HPRD,BioGRID | 11113190 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | ETO interacts with N-CoR. | BIND | 15333839 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD,BioGRID | 11113190 |
| NECAB2 | EFCBP2 | N-terminal EF-hand calcium binding protein 2 | Two-hybrid | BioGRID | 16189514 |
| OTUD4 | DKFZp434I0721 | DUBA6 | HIN1 | HSHIN1 | KIAA1046 | OTU domain containing 4 | Affinity Capture-MS | BioGRID | 17353931 |
| PRKAR2A | MGC3606 | PKR2 | PRKAR2 | protein kinase, cAMP-dependent, regulatory, type II, alpha | - | HPRD,BioGRID | 11593431 |
| RUNX1T1 | AML1T1 | CBFA2T1 | CDR | ETO | MGC2796 | MTG8 | MTG8b | ZMYND2 | runt-related transcription factor 1; translocated to, 1 (cyclin D-related) | - | HPRD,BioGRID | 11113190 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | - | HPRD,BioGRID | 11150306 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | ETO interacts with Sin3A. | BIND | 15333839 |
| TAF9B | DN-7 | DN7 | TAF9L | TAFII31L | TFIID-31 | TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa | Two-hybrid | BioGRID | 16189514 |
| TCF12 | HEB | HTF4 | HsT17266 | bHLHb20 | transcription factor 12 | ETO interacts with HEBa. This interaction was modeled on a demonstrated interaction between ETO and HEBb. | BIND | 15333839 |
| TCF12 | HEB | HTF4 | HsT17266 | bHLHb20 | transcription factor 12 | ETO interacts with HEBb. | BIND | 15333839 |
| TCF3 | E2A | ITF1 | MGC129647 | MGC129648 | bHLHb21 | transcription factor 3 (E2A immunoglobulin enhancer binding factors E12/E47) | ETO interacts with E2A. | BIND | 15333839 |
| TCF4 | E2-2 | ITF2 | MGC149723 | MGC149724 | PTHS | SEF2 | SEF2-1 | SEF2-1A | SEF2-1B | bHLHb19 | transcription factor 4 | ETO interacts with E2-2. | BIND | 15333839 |
| THRAP3 | FLJ22082 | MGC133082 | MGC133083 | TRAP150 | thyroid hormone receptor associated protein 3 | Affinity Capture-MS | BioGRID | 17353931 |
| VDR | NR1I1 | vitamin D (1,25- dihydroxyvitamin D3) receptor | Affinity Capture-Western Reconstituted Complex | BioGRID | 12460926 |
| WBP11 | DKFZp779M1063 | NPWBP | SIPP1 | WW domain binding protein 11 | Two-hybrid | BioGRID | 16189514 |
| ZBTB16 | PLZF | ZNF145 | zinc finger and BTB domain containing 16 | - | HPRD,BioGRID | 10688654 |11090081 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG ACUTE MYELOID LEUKEMIA | 60 | 47 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS BLUE DN | 61 | 35 | All SZGR 2.0 genes in this pathway |
| GARGALOVIC RESPONSE TO OXIDIZED PHOSPHOLIPIDS GREY DN | 74 | 44 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 MUTATED SIGNATURE 1 DN | 126 | 86 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 DN | 281 | 186 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN NPM1 SIGNATURE 3 DN | 162 | 116 | All SZGR 2.0 genes in this pathway |
| LEE NEURAL CREST STEM CELL UP | 146 | 99 | All SZGR 2.0 genes in this pathway |
| WAMUNYOKOLI OVARIAN CANCER LMP DN | 199 | 124 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| DUNNE TARGETS OF AML1 MTG8 FUSION DN | 19 | 17 | All SZGR 2.0 genes in this pathway |
| WONG ENDMETRIUM CANCER DN | 82 | 53 | All SZGR 2.0 genes in this pathway |
| RIZ ERYTHROID DIFFERENTIATION HEMGN | 31 | 21 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| DAWSON METHYLATED IN LYMPHOMA TCL1 | 59 | 45 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC | 35 | 25 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 8Q12 Q22 AMPLICON | 132 | 82 | All SZGR 2.0 genes in this pathway |
| ROSS ACUTE MYELOID LEUKEMIA CBF | 82 | 57 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS PLASMABLAST UP | 398 | 262 | All SZGR 2.0 genes in this pathway |
| WONG IFNA2 RESISTANCE DN | 34 | 20 | All SZGR 2.0 genes in this pathway |
| ROSS AML WITH AML1 ETO FUSION | 76 | 55 | All SZGR 2.0 genes in this pathway |
| BASSO CD40 SIGNALING DN | 68 | 44 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR DN | 101 | 70 | All SZGR 2.0 genes in this pathway |
| MARIADASON REGULATED BY HISTONE ACETYLATION DN | 54 | 30 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS PEAK AT 0HR | 63 | 48 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR DN | 101 | 64 | All SZGR 2.0 genes in this pathway |
| KONDO PROSTATE CANCER WITH H3K27ME3 | 196 | 93 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER POOR SURVIVAL UP | 31 | 22 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| YAUCH HEDGEHOG SIGNALING PARACRINE DN | 264 | 159 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| ZHANG TLX TARGETS 60HR UP | 293 | 203 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 13 | 30 | 20 | All SZGR 2.0 genes in this pathway |
| VALK AML WITH T 8 21 TRANSLOCATION | 5 | 5 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 4HR DN | 254 | 158 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS GROWING | 243 | 155 | All SZGR 2.0 genes in this pathway |
| CHICAS RB1 TARGETS CONFLUENT | 567 | 365 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 TARGETS 10HR DN | 244 | 157 | All SZGR 2.0 genes in this pathway |
| RATTENBACHER BOUND BY CELF1 | 467 | 251 | All SZGR 2.0 genes in this pathway |
| RAO BOUND BY SALL4 ISOFORM A | 182 | 108 | All SZGR 2.0 genes in this pathway |