Gene Page: CDK13
Summary ?
| GeneID | 8621 |
| Symbol | CDK13 |
| Synonyms | CDC2L|CDC2L5|CHED|hCDK13 |
| Description | cyclin-dependent kinase 13 |
| Reference | MIM:603309|HGNC:HGNC:1733|Ensembl:ENSG00000065883|HPRD:04496|Vega:OTTHUMG00000023726 |
| Gene type | protein-coding |
| Map location | 7p13 |
| Pascal p-value | 0.607 |
| Sherlock p-value | 0.659 |
| Fetal beta | 1.727 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 2 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg02853322 | 7 | 39989723 | CDK13 | 4.744E-4 | -0.317 | 0.046 | DMG:Wockner_2014 |
| cg09470376 | 7 | 39991217 | CDK13 | 5.62E-9 | 0.007 | 3.08E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
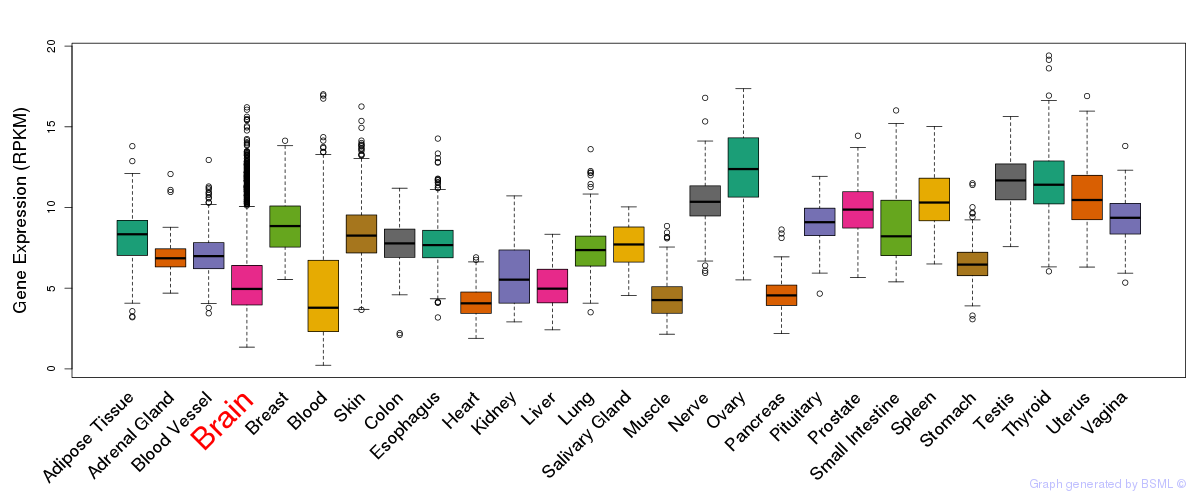
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| USP47 | 0.95 | 0.96 |
| TOPORS | 0.95 | 0.94 |
| CTR9 | 0.94 | 0.95 |
| NOL8 | 0.94 | 0.94 |
| ZNF644 | 0.94 | 0.95 |
| SR140 | 0.94 | 0.94 |
| SUPT16H | 0.93 | 0.95 |
| ZNF225 | 0.93 | 0.93 |
| ZNF280D | 0.93 | 0.92 |
| SIRT1 | 0.93 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.75 | -0.86 |
| MT-CO2 | -0.74 | -0.85 |
| IFI27 | -0.74 | -0.86 |
| FXYD1 | -0.74 | -0.84 |
| AF347015.27 | -0.73 | -0.82 |
| AF347015.33 | -0.71 | -0.81 |
| HIGD1B | -0.71 | -0.83 |
| MT-CYB | -0.71 | -0.81 |
| AF347015.8 | -0.70 | -0.83 |
| HLA-F | -0.70 | -0.74 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004693 | cyclin-dependent protein kinase activity | IEA | neuron (GO term level: 8) | - |
| GO:0000166 | nucleotide binding | IEA | - | |
| GO:0005524 | ATP binding | IEA | - | |
| GO:0004674 | protein serine/threonine kinase activity | IEA | - | |
| GO:0016740 | transferase activity | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006468 | protein amino acid phosphorylation | IEA | - | |
| GO:0007088 | regulation of mitosis | TAS | 1731328 | |
| GO:0008284 | positive regulation of cell proliferation | TAS | 1731328 | |
| GO:0007275 | multicellular organismal development | TAS | 1731328 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN | 463 | 290 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| NUNODA RESPONSE TO DASATINIB IMATINIB UP | 29 | 20 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| HEIDENBLAD AMPLICON 12P11 12 DN | 30 | 16 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| LIU COMMON CANCER GENES | 79 | 47 | All SZGR 2.0 genes in this pathway |
| LIAO METASTASIS | 539 | 324 | All SZGR 2.0 genes in this pathway |
| SCHWAB TARGETS OF BMYB POLYMORPHIC VARIANTS DN | 17 | 12 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| HADDAD B LYMPHOCYTE PROGENITOR | 293 | 193 | All SZGR 2.0 genes in this pathway |
| KANG IMMORTALIZED BY TERT UP | 89 | 61 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| NATSUME RESPONSE TO INTERFERON BETA UP | 71 | 49 | All SZGR 2.0 genes in this pathway |
| KAAB HEART ATRIUM VS VENTRICLE UP | 249 | 170 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR DN | 504 | 323 | All SZGR 2.0 genes in this pathway |
| MONNIER POSTRADIATION TUMOR ESCAPE DN | 373 | 196 | All SZGR 2.0 genes in this pathway |
| BLUM RESPONSE TO SALIRASIB UP | 245 | 159 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED UP | 85 | 50 | All SZGR 2.0 genes in this pathway |
| STAMBOLSKY TARGETS OF MUTATED TP53 UP | 49 | 35 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 2 DN | 336 | 211 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-133 | 348 | 354 | 1A | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-141/200a | 488 | 495 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-148/152 | 410 | 416 | 1A | hsa-miR-148a | UCAGUGCACUACAGAACUUUGU |
| hsa-miR-152brain | UCAGUGCAUGACAGAACUUGGG | ||||
| hsa-miR-148b | UCAGUGCAUCACAGAACUUUGU | ||||
| miR-153 | 317 | 323 | m8 | hsa-miR-153 | UUGCAUAGUCACAAAAGUGA |
| miR-19 | 474 | 480 | m8 | hsa-miR-19a | UGUGCAAAUCUAUGCAAAACUGA |
| hsa-miR-19b | UGUGCAAAUCCAUGCAAAACUGA | ||||
| miR-26 | 79 | 85 | 1A | hsa-miR-26abrain | UUCAAGUAAUCCAGGAUAGGC |
| hsa-miR-26bSZ | UUCAAGUAAUUCAGGAUAGGUU | ||||
| miR-320 | 103 | 110 | 1A,m8 | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-335 | 546 | 552 | 1A | hsa-miR-335brain | UCAAGAGCAAUAACGAAAAAUGU |
| miR-455 | 18 | 24 | m8 | hsa-miR-455 | UAUGUGCCUUUGGACUACAUCG |
| miR-488 | 31 | 38 | 1A,m8 | hsa-miR-488 | CCCAGAUAAUGGCACUCUCAA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.