Gene Page: RUNX3
Summary ?
| GeneID | 864 |
| Symbol | RUNX3 |
| Synonyms | AML2|CBFA3|PEBP2aC |
| Description | runt related transcription factor 3 |
| Reference | MIM:600210|HGNC:HGNC:10473|Ensembl:ENSG00000020633|HPRD:02565|Vega:OTTHUMG00000003316 |
| Gene type | protein-coding |
| Map location | 1p36 |
| Pascal p-value | 0.317 |
| TADA p-value | 0.012 |
| Fetal beta | 0.053 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 2 |
| DNM:Gulsuner_2013 | Whole Exome Sequencing analysis | 155 DNMs identified by exome sequencing of quads or trios of schizophrenia individuals and their parents. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| RUNX3 | chr1 | 25233863 | C | T | NM_001031680 NM_004350 | p.211R>H p.197R>H | missense missense | Schizophrenia | DNM:Gulsuner_2013 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06037693 | 1 | 25292072 | RUNX3 | 7.24E-8 | -8.177 | DMG:vanEijk_2014 | |
| cg16529592 | 1 | 25292215 | RUNX3 | 2E-6 | -8.26 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
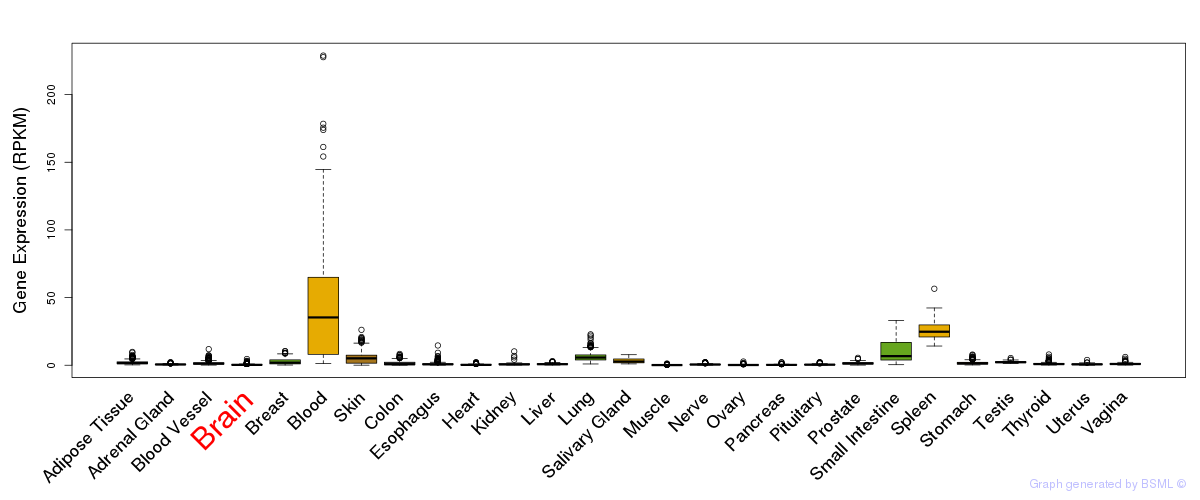
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRCH1 | 0.94 | 0.92 |
| SSBP2 | 0.94 | 0.89 |
| EIF4B | 0.94 | 0.91 |
| SEMA3A | 0.93 | 0.91 |
| USP3 | 0.93 | 0.88 |
| SLAIN2 | 0.92 | 0.93 |
| XRN2 | 0.92 | 0.95 |
| PRRC1 | 0.92 | 0.93 |
| KDM5B | 0.92 | 0.95 |
| C9orf97 | 0.92 | 0.93 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LHPP | -0.66 | -0.66 |
| TNFSF12 | -0.65 | -0.71 |
| C5orf53 | -0.65 | -0.81 |
| FBXO2 | -0.65 | -0.70 |
| HLA-F | -0.64 | -0.83 |
| SERPINB6 | -0.64 | -0.74 |
| AIFM3 | -0.64 | -0.82 |
| SLC16A11 | -0.63 | -0.70 |
| ALDOC | -0.63 | -0.74 |
| PTH1R | -0.63 | -0.76 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CBFB | PEBP2B | core-binding factor, beta subunit | - | HPRD | 7622058 |
| CBFB | PEBP2B | core-binding factor, beta subunit | Cbf-beta interacts with AML2. This interaction was modeled on a demonstrated interaction between mouse Cbf-beta and human AML2. | BIND | 9751710 |
| EP300 | KAT3B | p300 | E1A binding protein p300 | - | HPRD | 15138260 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD | 15138260 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | - | HPRD | 15138260 |
| HDAC4 | HA6116 | HD4 | HDAC-A | HDACA | KIAA0288 | histone deacetylase 4 | - | HPRD | 15138260 |
| HDAC5 | FLJ90614 | HD5 | NY-CO-9 | histone deacetylase 5 | - | HPRD | 15138260 |
| PHB2 | BAP | BCAP37 | Bap37 | MGC117268 | PNAS-141 | REA | p22 | prohibitin 2 | Affinity Capture-MS | BioGRID | 17353931 |
| SMAD1 | BSP1 | JV4-1 | JV41 | MADH1 | MADR1 | SMAD family member 1 | Affinity Capture-Western | BioGRID | 10531362 |
| SMAD2 | JV18 | JV18-1 | MADH2 | MADR2 | MGC22139 | MGC34440 | hMAD-2 | hSMAD2 | SMAD family member 2 | Affinity Capture-Western | BioGRID | 10531362 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | - | HPRD | 15138260 |
| SMAD3 | DKFZp586N0721 | DKFZp686J10186 | HSPC193 | HsT17436 | JV15-2 | MADH3 | MGC60396 | SMAD family member 3 | Affinity Capture-Western | BioGRID | 10531362 |
| SMAD5 | DKFZp781C1895 | DKFZp781O1323 | Dwfc | JV5-1 | MADH5 | SMAD family member 5 | Affinity Capture-Western | BioGRID | 10531362 |
| SMURF1 | KIAA1625 | SMAD specific E3 ubiquitin protein ligase 1 | - | HPRD | 15138260 |
| SMURF2 | DKFZp686F0270 | MGC138150 | SMAD specific E3 ubiquitin protein ligase 2 | - | HPRD | 15138260 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | AML2 interacts with TLE1. | BIND | 9751710 |
| TLE1 | ESG | ESG1 | GRG1 | transducin-like enhancer of split 1 (E(sp1) homolog, Drosophila) | - | HPRD,BioGRID | 9751710 |
| YAP1 | YAP | YAP2 | YAP65 | YKI | Yes-associated protein 1, 65kDa | Reconstituted Complex | BioGRID | 10228168 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 UP | 201 | 125 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP | 473 | 314 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL DN | 455 | 304 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| HOEBEKE LYMPHOID STEM CELL UP | 95 | 64 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION HSC DN | 187 | 115 | All SZGR 2.0 genes in this pathway |
| CHOW RASSF1 TARGETS DN | 29 | 19 | All SZGR 2.0 genes in this pathway |
| GAUSSMANN MLL AF4 FUSION TARGETS B UP | 26 | 16 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| MUELLER METHYLATED IN GLIOBLASTOMA | 40 | 22 | All SZGR 2.0 genes in this pathway |
| PEREZ TP63 TARGETS | 355 | 243 | All SZGR 2.0 genes in this pathway |
| MARTIN INTERACT WITH HDAC | 44 | 31 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| DIRMEIER LMP1 RESPONSE LATE UP | 57 | 41 | All SZGR 2.0 genes in this pathway |
| AMUNDSON RESPONSE TO ARSENITE | 217 | 143 | All SZGR 2.0 genes in this pathway |
| LI WILMS TUMOR VS FETAL KIDNEY 2 DN | 51 | 42 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 60 HELA | 46 | 32 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 120 HELA | 69 | 47 | All SZGR 2.0 genes in this pathway |
| ZHANG TARGETS OF EWSR1 FLI1 FUSION | 88 | 68 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| VISALA AGING LYMPHOCYTE DN | 19 | 10 | All SZGR 2.0 genes in this pathway |
| RODWELL AGING KIDNEY UP | 487 | 303 | All SZGR 2.0 genes in this pathway |
| BILD E2F3 ONCOGENIC SIGNATURE | 246 | 153 | All SZGR 2.0 genes in this pathway |
| LOPES METHYLATED IN COLON CANCER DN | 28 | 26 | All SZGR 2.0 genes in this pathway |
| WALLACE PROSTATE CANCER RACE UP | 299 | 167 | All SZGR 2.0 genes in this pathway |
| CHENG IMPRINTED BY ESTRADIOL | 110 | 68 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| HOQUE METHYLATED IN CANCER | 56 | 45 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER RELAPSE IN BONE DN | 315 | 197 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL UP | 648 | 398 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| FINAK BREAST CANCER SDPP SIGNATURE | 26 | 11 | All SZGR 2.0 genes in this pathway |
| ZHAN EARLY DIFFERENTIATION GENES DN | 42 | 29 | All SZGR 2.0 genes in this pathway |
| VALK AML CLUSTER 9 | 35 | 26 | All SZGR 2.0 genes in this pathway |
| JISON SICKLE CELL DISEASE DN | 181 | 97 | All SZGR 2.0 genes in this pathway |
| ONO AML1 TARGETS DN | 41 | 25 | All SZGR 2.0 genes in this pathway |
| ONO FOXP3 TARGETS DN | 42 | 23 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA DN | 80 | 53 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP D | 280 | 158 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| ALFANO MYC TARGETS | 239 | 156 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |