Gene Page: CBL
Summary ?
| GeneID | 867 |
| Symbol | CBL |
| Synonyms | C-CBL|CBL2|FRA11B|NSLL|RNF55 |
| Description | Cbl proto-oncogene, E3 ubiquitin protein ligase |
| Reference | MIM:165360|HGNC:HGNC:1541|Ensembl:ENSG00000110395|HPRD:01320|Vega:OTTHUMG00000166170 |
| Gene type | protein-coding |
| Map location | 11q23.3 |
| Pascal p-value | 0.141 |
| Fetal beta | 0.019 |
| eGene | Cerebellum |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| GSMA_I | Genome scan meta-analysis | Psr: 0.006 | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0043 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
General gene expression (GTEx)
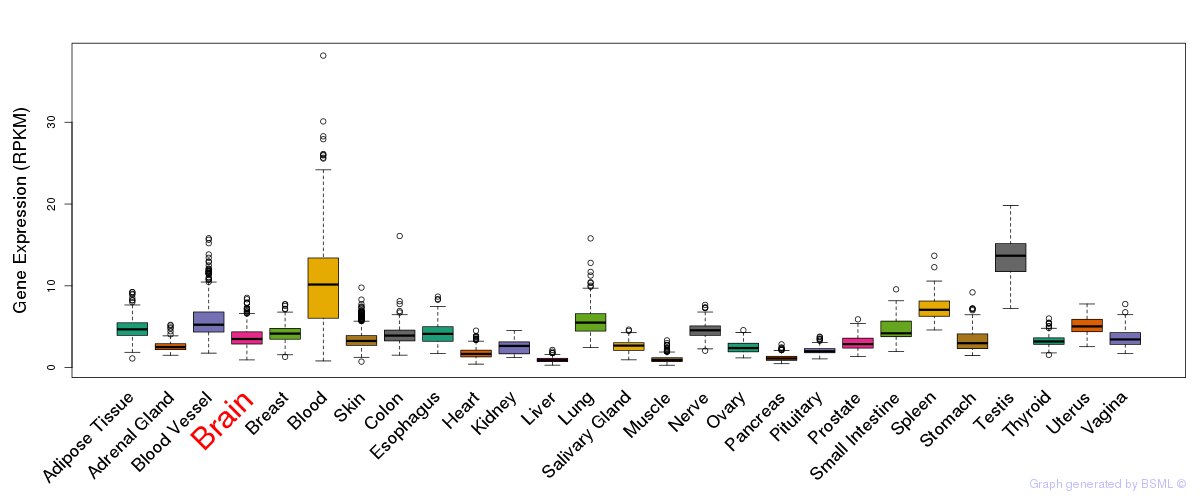
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| STRA6 | 0.64 | 0.66 |
| MMP2 | 0.63 | 0.71 |
| EPHB4 | 0.63 | 0.71 |
| SLC3A2 | 0.63 | 0.64 |
| PDIA4 | 0.62 | 0.62 |
| HTRA3 | 0.61 | 0.68 |
| VASP | 0.60 | 0.72 |
| FSTL1 | 0.60 | 0.67 |
| NID1 | 0.60 | 0.74 |
| CMTM3 | 0.60 | 0.66 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| ASPA | -0.42 | -0.52 |
| RBP7 | -0.41 | -0.56 |
| OPALIN | -0.41 | -0.56 |
| EVI2A | -0.40 | -0.52 |
| MAL | -0.40 | -0.49 |
| RNASE1 | -0.40 | -0.47 |
| CAPN3 | -0.39 | -0.57 |
| SEPT4 | -0.39 | -0.50 |
| DBNDD2 | -0.39 | -0.45 |
| PLLP | -0.39 | -0.46 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004871 | signal transducer activity | IEA | - | |
| GO:0003700 | transcription factor activity | TAS | 2030914 | |
| GO:0005509 | calcium ion binding | IEA | - | |
| GO:0005515 | protein binding | IPI | 11891219 | |
| GO:0005525 | GTP binding | IEA | - | |
| GO:0004842 | ubiquitin-protein ligase activity | EXP | 12218189 |12593796 |16407834 | |
| GO:0004842 | ubiquitin-protein ligase activity | TAS | 15465819 | |
| GO:0016874 | ligase activity | IEA | - | |
| GO:0008270 | zinc ion binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007173 | epidermal growth factor receptor signaling pathway | TAS | 15465819 | |
| GO:0006511 | ubiquitin-dependent protein catabolic process | IEA | - | |
| GO:0007166 | cell surface receptor linked signal transduction | TAS | 15465819 | |
| GO:0007017 | microtubule-based process | IEA | - | |
| GO:0016567 | protein ubiquitination | TAS | 15465819 | |
| GO:0048260 | positive regulation of receptor-mediated endocytosis | TAS | 15465819 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005829 | cytosol | EXP | 8621719 |11823423 |14505571 | |
| GO:0005874 | microtubule | IEA | - | |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 10567358 |11894095 |12218189 |12593796 |15475003 |15962011 |16407834 | |
| GO:0005886 | plasma membrane | IDA | 15465819 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABL1 | ABL | JTK7 | bcr/abl | c-ABL | p150 | v-abl | c-abl oncogene 1, receptor tyrosine kinase | - | HPRD,BioGRID | 12475393 |
| ARHGEF7 | BETA-PIX | COOL1 | DKFZp686C12170 | DKFZp761K1021 | KIAA0142 | KIAA0412 | Nbla10314 | P50 | P50BP | P85 | P85COOL1 | P85SPR | PAK3 | PIXB | Rho guanine nucleotide exchange factor (GEF) 7 | Two-hybrid | BioGRID | 12935897 |
| BCR | ALL | BCR-ABL1 | BCR1 | CML | D22S11 | D22S662 | FLJ16453 | PHL | breakpoint cluster region | Co-purification | BioGRID | 8641358 |
| BLK | MGC10442 | B lymphoid tyrosine kinase | - | HPRD | 8083187 |
| BTK | AGMX1 | AT | ATK | BPK | IMD1 | MGC126261 | MGC126262 | PSCTK1 | XLA | Bruton agammaglobulinemia tyrosine kinase | - | HPRD,BioGRID | 7629518 |10427990 |
| CD19 | B4 | MGC12802 | CD19 molecule | - | HPRD | 9178909 |
| CD2AP | CMS | DKFZp586H0519 | CD2-associated protein | CMS interacts with Cbl. This interaction was modeled on a demonstrated interaction between CMS from an unspecified species and human Cbl. | BIND | 15128873 |
| CD2AP | CMS | DKFZp586H0519 | CD2-associated protein | - | HPRD,BioGRID | 11067845 |
| CD5 | LEU1 | T1 | CD5 molecule | - | HPRD,BioGRID | 9603468 |
| CDKL2 | KKIAMRE | P56 | cyclin-dependent kinase-like 2 (CDC2-related kinase) | - | HPRD | 7721825 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | Affinity Capture-Western | BioGRID | 9178909 |9461587 |
| CRK | CRKII | v-crk sarcoma virus CT10 oncogene homolog (avian) | - | HPRD | 8621483 |8662998 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | Affinity Capture-Western Reconstituted Complex | BioGRID | 9092574 |9162067 |9344843 |9461587 |9590251 |9820532 |10204582 |10907644 |11133830 |12244174 |
| CRKL | - | v-crk sarcoma virus CT10 oncogene homolog (avian)-like | - | HPRD | 9162067 |9590251 |12244174 |
| CSF1R | C-FMS | CD115 | CSFR | FIM2 | FMS | colony stimulating factor 1 receptor | - | HPRD,BioGRID | 11847211 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD,BioGRID | 11149930 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | Affinity Capture-Western | BioGRID | 10086340 |12061819 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | - | HPRD | 8662998 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | An unspecified isoform of EGFR interacts with CBL. | BIND | 15735736 |
| EGFR | ERBB | ERBB1 | HER1 | PIG61 | mENA | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | c-Cbl ubiquitinates an unspecified isoform of EGFR. This interaction was modeled on a demonstrated interaction between c-Cbl and EGFR, both from an unspecified species. | BIND | 12234920 |
| EPHB6 | HEP | MGC129910 | MGC129911 | EPH receptor B6 | - | HPRD | 11713248 |
| FGR | FLJ43153 | MGC75096 | SRC2 | c-fgr | c-src2 | p55c-fgr | p58c-fgr | Gardner-Rasheed feline sarcoma viral (v-fgr) oncogene homolog | - | HPRD,BioGRID | 12435267 |
| FLOT1 | - | flotillin 1 | - | HPRD,BioGRID | 11001060 |
| FRS2 | FRS2A | FRS2alpha | SNT | SNT-1 | SNT1 | fibroblast growth factor receptor substrate 2 | - | HPRD,BioGRID | 11997436 |
| FYN | MGC45350 | SLK | SYN | FYN oncogene related to SRC, FGR, YES | - | HPRD,BioGRID | 9535867 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Grb2 interacts with Cbl. | BIND | 15782196 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | c-Cbl interacts with Grb2. This interaction was modeled on a demonstrated interaction between human c-Cbl and Grb2 from an unspecified species. | BIND | 11053437 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 8083187 |9108392 |9174058 |9178909 |9367879 |9461587 |9872323 |10086340 |10204582 |10570290 |11964172 |11997436 |
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD | 9108392 |9178909 |11133830 |11964172 |
| HCK | JTK9 | hemopoietic cell kinase | - | HPRD,BioGRID | 10092522 |
| INPPL1 | SHIP2 | inositol polyphosphate phosphatase-like 1 | - | HPRD,BioGRID | 12504111 |
| ITK | EMT | LYK | MGC126257 | MGC126258 | PSCTK2 | IL2-inducible T-cell kinase | - | HPRD,BioGRID | 8810341 |
| KRT18 | CYK18 | K18 | keratin 18 | c-Cbl interacts with Keratin 18. | BIND | 9367879 |
| KRT18 | CYK18 | K18 | keratin 18 | - | HPRD,BioGRID | 9367879 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD,BioGRID | 9178909 |11741956 |11904433 |
| LCP2 | SLP-76 | SLP76 | lymphocyte cytosolic protein 2 (SH2 domain containing leukocyte protein of 76kDa) | - | HPRD,BioGRID | 10204582 |
| LTK | TYK1 | leukocyte receptor tyrosine kinase | - | HPRD,BioGRID | 9223670 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | Affinity Capture-Western | BioGRID | 12244174 |
| LYN | FLJ26625 | JTK8 | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | - | HPRD | 12149655 |
| MET | AUTS9 | HGFR | RCCP2 | c-Met | met proto-oncogene (hepatocyte growth factor receptor) | Met interacts with Cbl. | BIND | 11894096 |
| MET | AUTS9 | HGFR | RCCP2 | c-Met | met proto-oncogene (hepatocyte growth factor receptor) | - | HPRD,BioGRID | 11894096 |
| MYO1C | FLJ23903 | MMI-beta | MMIb | NMI | myr2 | myosin IC | Two-hybrid | BioGRID | 9367879 |
| NCK1 | MGC12668 | NCK | NCKalpha | NCK adaptor protein 1 | Affinity Capture-Western | BioGRID | 10204582 |11494134 |
| OSTF1 | FLJ20559 | OSF | SH3P2 | bA235O14.1 | osteoclast stimulating factor 1 | Cbl interacts with SH3P2. | BIND | 15135048 |
| PDGFRA | CD140A | MGC74795 | PDGFR2 | Rhe-PDGFRA | platelet-derived growth factor receptor, alpha polypeptide | Affinity Capture-Western Reconstituted Complex | BioGRID | 9234717 |
| PDGFRB | CD140B | JTK12 | PDGF-R-beta | PDGFR | PDGFR1 | platelet-derived growth factor receptor, beta polypeptide | c-Cbl interacts with and ubiquitinates PDGF-R-beta. This interaction was modeled on a demonstrated interaction between human c-Cbl and PDGF-R-beta from an unspecified species. | BIND | 10514377 |
| PIK3CA | MGC142161 | MGC142163 | PI3K | p110-alpha | phosphoinositide-3-kinase, catalytic, alpha polypeptide | Co-purification | BioGRID | 8641358 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | Affinity Capture-Western | BioGRID | 9461587 |9918857 |
| PIK3R2 | P85B | p85 | p85-BETA | phosphoinositide-3-kinase, regulatory subunit 2 (beta) | - | HPRD,BioGRID | 7629144 |
| PLCG1 | PLC-II | PLC1 | PLC148 | PLCgamma1 | phospholipase C, gamma 1 | - | HPRD,BioGRID | 9712732 |12061819 |
| PTK2B | CADTK | CAKB | FADK2 | FAK2 | FRNK | PKB | PTK | PYK2 | RAFTK | PTK2B protein tyrosine kinase 2 beta | - | HPRD,BioGRID | 11149930 |
| PTPN22 | LYP | Lyp1 | Lyp2 | PEP | PTPN8 | protein tyrosine phosphatase, non-receptor type 22 (lymphoid) | - | HPRD | 10068674 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 12176909 |
| PXN | FLJ16691 | paxillin | Co-purification | BioGRID | 8641358 |
| RET | CDHF12 | HSCR1 | MEN2A | MEN2B | MTC1 | PTC | RET-ELE1 | RET51 | ret proto-oncogene | - | HPRD | 12727845 |
| SH2B2 | APS | SH2B adaptor protein 2 | Reconstituted Complex | BioGRID | 10374881 |
| SH3BP2 | 3BP2 | CRBM | CRPM | FLJ42079 | RES4-23 | SH3-domain binding protein 2 | Reconstituted Complex | BioGRID | 9846481 |
| SH3GL2 | CNSA2 | EEN-B1 | FLJ20276 | FLJ25015 | SH3D2A | SH3P4 | SH3-domain GRB2-like 2 | - | HPRD | 11894096 |
| SH3GL2 | CNSA2 | EEN-B1 | FLJ20276 | FLJ25015 | SH3D2A | SH3P4 | SH3-domain GRB2-like 2 | Affinity Capture-Western | BioGRID | 11894095 |
| SH3GL3 | CNSA3 | EEN-2B-L3 | EEN-B2 | HsT19371 | SH3D2C | SH3P13 | SH3-domain GRB2-like 3 | - | HPRD,BioGRID | 11894096 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | - | HPRD,BioGRID | 11894095 |
| SH3KBP1 | CIN85 | GIG10 | MIG18 | SH3-domain kinase binding protein 1 | CIN85 interacts with Cbl. | BIND | 11894095 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD,BioGRID | 10570290 |
| SLA | SLA1 | SLAP | Src-like-adaptor | - | HPRD,BioGRID | 10449770 |
| SLA2 | C20orf156 | FLJ21992 | MGC49845 | SLAP-2 | SLAP2 | Src-like-adaptor 2 | - | HPRD,BioGRID | 11891219 |12024036 |
| SORBS1 | CAP | DKFZp451C066 | DKFZp586P1422 | FLAF2 | FLJ12406 | KIAA1296 | R85FL | SH3D5 | SH3P12 | SORB1 | sorbin and SH3 domain containing 1 | - | HPRD | 11001060 |
| SORBS1 | CAP | DKFZp451C066 | DKFZp586P1422 | FLAF2 | FLJ12406 | KIAA1296 | R85FL | SH3D5 | SH3P12 | SORB1 | sorbin and SH3 domain containing 1 | Affinity Capture-Western | BioGRID | 12504111 |15128873 |
| SORBS1 | CAP | DKFZp451C066 | DKFZp586P1422 | FLAF2 | FLJ12406 | KIAA1296 | R85FL | SH3D5 | SH3P12 | SORB1 | sorbin and SH3 domain containing 1 | An unspecified isoform of CAP interacts with Cbl. This interaction was modeled on a demonstrated interaction between CAP from an unspecified species and human Cbl. | BIND | 15128873 |
| SORBS2 | ARGBP2 | FLJ93447 | KIAA0777 | PRO0618 | sorbin and SH3 domain containing 2 | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 12475393 |15128873 |
| SORBS2 | ARGBP2 | FLJ93447 | KIAA0777 | PRO0618 | sorbin and SH3 domain containing 2 | ArgBP2 interacts with Cbl. | BIND | 15128873 |
| SORBS2 | ARGBP2 | FLJ93447 | KIAA0777 | PRO0618 | sorbin and SH3 domain containing 2 | ArgBP2-A interacts with Cbl. | BIND | 12475393 |
| SPRY2 | MGC23039 | hSPRY2 | sprouty homolog 2 (Drosophila) | - | HPRD,BioGRID | 11053437 |12234920 |
| SPRY2 | MGC23039 | hSPRY2 | sprouty homolog 2 (Drosophila) | c-Cbl interacts with hSpry2. This interaction was modeled on a demonstrated interaction between c-Cbl from an unspecified species and human hSpry2. | BIND | 12234920 |
| SPRY2 | MGC23039 | hSPRY2 | sprouty homolog 2 (Drosophila) | SPRY2 interacts with c-Cbl. | BIND | 11053437 |
| SPRY2 | MGC23039 | hSPRY2 | sprouty homolog 2 (Drosophila) | hSpry2 interacts with c-Cbl. | BIND | 12815057 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | - | HPRD | 8635998 |
| SRC | ASV | SRC1 | c-SRC | p60-Src | v-src sarcoma (Schmidt-Ruppin A-2) viral oncogene homolog (avian) | Cbl interacts with Src. This interaction was modeled on a demonstrated interaction between human Cbl and Src from unspecified species. | BIND | 15135048 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | Affinity Capture-Western Reconstituted Complex | BioGRID | 9857068 |11331248 |12435267 |
| SYK | DKFZp313N1010 | FLJ25043 | FLJ37489 | spleen tyrosine kinase | - | HPRD | 9857068 |10540342 |11133830 |
| TLN1 | ILWEQ | KIAA1027 | TLN | talin 1 | Co-purification | BioGRID | 8641358 |
| TYK2 | JTK1 | tyrosine kinase 2 | Affinity Capture-Western | BioGRID | 8780698 |
| UBASH3B | KIAA1959 | MGC15437 | STS-1 | STS1 | p70 | ubiquitin associated and SH3 domain containing, B | - | HPRD | 15159412 |
| UBE2D2 | E2(17)KB2 | PUBC1 | UBC4 | UBC4/5 | UBCH5B | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | - | HPRD,BioGRID | 10514377 |
| UBE2D2 | E2(17)KB2 | PUBC1 | UBC4 | UBC4/5 | UBCH5B | ubiquitin-conjugating enzyme E2D 2 (UBC4/5 homolog, yeast) | c-Cbl interacts with Ubc4. | BIND | 10514377 |
| UBE2G1 | E217K | UBC7 | UBE2G | ubiquitin-conjugating enzyme E2G 1 (UBC7 homolog, yeast) | - | HPRD | 11032556 |
| UBE2G2 | UBC7 | ubiquitin-conjugating enzyme E2G 2 (UBC7 homolog, yeast) | - | HPRD | 11032556 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | Affinity Capture-Western Co-crystal Structure Reconstituted Complex | BioGRID | 10531381 |10966114 |12234920 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | - | HPRD | 10531381 |10966114 |11278816 |
| UBE2L3 | E2-F1 | L-UBC | UBCH7 | UbcM4 | ubiquitin-conjugating enzyme E2L 3 | c-Cbl interacts with an unspecified isoform of UbcH7. This interaction was modeled on a demonstrated interaction between c-Cbl from an unspecified species and human UbcH7. | BIND | 12234920 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | - | HPRD | 9200440 |11133830 |
| VAV1 | VAV | vav 1 guanine nucleotide exchange factor | Affinity Capture-Western Far Western Reconstituted Complex | BioGRID | 9200440 |11331248 |
| VAV2 | - | vav 2 guanine nucleotide exchange factor | - | HPRD | 11313921 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | c-Cbl interacts with 14-3-3-beta. | BIND | 9367879 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9367879 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | - | HPRD,BioGRID | 8663231 |11024037 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Two-hybrid | BioGRID | 9367879 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | c-Cbl interacts with 14-3-3-zeta. | BIND | 9367879 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | Affinity Capture-Western Co-crystal Structure | BioGRID | 8798643 |10078535 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ERBB SIGNALING PATHWAY | 87 | 71 | All SZGR 2.0 genes in this pathway |
| KEGG UBIQUITIN MEDIATED PROTEOLYSIS | 138 | 98 | All SZGR 2.0 genes in this pathway |
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| KEGG JAK STAT SIGNALING PATHWAY | 155 | 105 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| KEGG CHRONIC MYELOID LEUKEMIA | 73 | 59 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CBL PATHWAY | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL2RB PATHWAY | 38 | 26 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SPRY PATHWAY | 18 | 15 | All SZGR 2.0 genes in this pathway |
| SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | 51 | 41 | All SZGR 2.0 genes in this pathway |
| ST T CELL SIGNAL TRANSDUCTION | 45 | 33 | All SZGR 2.0 genes in this pathway |
| PID FCER1 PATHWAY | 62 | 43 | All SZGR 2.0 genes in this pathway |
| PID INSULIN PATHWAY | 45 | 32 | All SZGR 2.0 genes in this pathway |
| PID NOTCH PATHWAY | 59 | 49 | All SZGR 2.0 genes in this pathway |
| PID IL4 2PATHWAY | 65 | 43 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID MET PATHWAY | 80 | 60 | All SZGR 2.0 genes in this pathway |
| PID REELIN PATHWAY | 29 | 29 | All SZGR 2.0 genes in this pathway |
| PID CDC42 PATHWAY | 70 | 51 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID AVB3 INTEGRIN PATHWAY | 75 | 53 | All SZGR 2.0 genes in this pathway |
| PID IFNG PATHWAY | 40 | 34 | All SZGR 2.0 genes in this pathway |
| PID EPHA FWDPATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID PDGFRB PATHWAY | 129 | 103 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR2 PATHWAY | 34 | 26 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 INTERNALIZATION PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 PATHWAY | 26 | 23 | All SZGR 2.0 genes in this pathway |
| PID KIT PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| PID EPO PATHWAY | 34 | 29 | All SZGR 2.0 genes in this pathway |
| PID VEGFR1 2 PATHWAY | 69 | 57 | All SZGR 2.0 genes in this pathway |
| PID IL8 CXCR1 PATHWAY | 28 | 19 | All SZGR 2.0 genes in this pathway |
| PID EPHA2 FWD PATHWAY | 19 | 16 | All SZGR 2.0 genes in this pathway |
| PID FGF PATHWAY | 55 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY SCF KIT | 78 | 59 | All SZGR 2.0 genes in this pathway |
| REACTOME SPRY REGULATION OF FGF SIGNALING | 14 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATION OF FGFR SIGNALING | 37 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF KIT SIGNALING | 17 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | 18 | 13 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY EGFR IN CANCER | 109 | 80 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIGEN ACTIVATES B CELL RECEPTOR LEADING TO GENERATION OF SECOND MESSENGERS | 29 | 24 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY THE B CELL RECEPTOR BCR | 126 | 90 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR IN DISEASE | 127 | 88 | All SZGR 2.0 genes in this pathway |
| REACTOME EGFR DOWNREGULATION | 25 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ILS | 107 | 86 | All SZGR 2.0 genes in this pathway |
| REACTOME REGULATION OF SIGNALING BY CBL | 18 | 16 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 3 5 AND GM CSF SIGNALING | 43 | 34 | All SZGR 2.0 genes in this pathway |
| REACTOME IL 6 SIGNALING | 11 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY FGFR | 112 | 80 | All SZGR 2.0 genes in this pathway |
| RODRIGUES NTN1 TARGETS DN | 158 | 102 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LASTOWSKA NEUROBLASTOMA COPY NUMBER DN | 800 | 473 | All SZGR 2.0 genes in this pathway |
| MYLLYKANGAS AMPLIFICATION HOT SPOT 23 | 24 | 13 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER DN | 514 | 319 | All SZGR 2.0 genes in this pathway |
| SHEN SMARCA2 TARGETS DN | 357 | 212 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| HEDENFALK BREAST CANCER BRCA1 VS BRCA2 | 163 | 113 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| GRESHOCK CANCER COPY NUMBER UP | 323 | 240 | All SZGR 2.0 genes in this pathway |
| LEE RECENT THYMIC EMIGRANT | 227 | 128 | All SZGR 2.0 genes in this pathway |
| FONTAINE PAPILLARY THYROID CARCINOMA DN | 80 | 53 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION DN | 321 | 200 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| PLASARI TGFB1 SIGNALING VIA NFIC 1HR DN | 106 | 77 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| let-7/98 | 3440 | 3447 | 1A,m8 | hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU |
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| hsa-let-7abrain | UGAGGUAGUAGGUUGUAUAGUU | ||||
| hsa-let-7bbrain | UGAGGUAGUAGGUUGUGUGGUU | ||||
| hsa-let-7cbrain | UGAGGUAGUAGGUUGUAUGGUU | ||||
| hsa-let-7dbrain | AGAGGUAGUAGGUUGCAUAGU | ||||
| hsa-let-7ebrain | UGAGGUAGGAGGUUGUAUAGU | ||||
| hsa-let-7fbrain | UGAGGUAGUAGAUUGUAUAGUU | ||||
| hsa-miR-98brain | UGAGGUAGUAAGUUGUAUUGUU | ||||
| hsa-let-7gSZ | UGAGGUAGUAGUUUGUACAGU | ||||
| hsa-let-7ibrain | UGAGGUAGUAGUUUGUGCUGU | ||||
| miR-1/206 | 914 | 921 | 1A,m8 | hsa-miR-1 | UGGAAUGUAAAGAAGUAUGUA |
| hsa-miR-206SZ | UGGAAUGUAAGGAAGUGUGUGG | ||||
| hsa-miR-613 | AGGAAUGUUCCUUCUUUGCC | ||||
| miR-101 | 763 | 769 | m8 | hsa-miR-101 | UACAGUACUGUGAUAACUGAAG |
| miR-124/506 | 7186 | 7192 | m8 | hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA |
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| hsa-miR-506 | UAAGGCACCCUUCUGAGUAGA | ||||
| hsa-miR-124brain | UAAGGCACGCGGUGAAUGCC | ||||
| miR-133 | 3362 | 3368 | m8 | hsa-miR-133a | UUGGUCCCCUUCAACCAGCUGU |
| hsa-miR-133b | UUGGUCCCCUUCAACCAGCUA | ||||
| miR-141/200a | 8015 | 8022 | 1A,m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-150 | 3871 | 3877 | 1A | hsa-miR-150 | UCUCCCAACCCUUGUACCAGUG |
| miR-192/215 | 985 | 991 | m8 | hsa-miR-192 | CUGACCUAUGAAUUGACAGCC |
| hsa-miR-215 | AUGACCUAUGAAUUGACAGAC | ||||
| miR-199 | 808 | 814 | m8 | hsa-miR-199a | CCCAGUGUUCAGACUACCUGUUC |
| hsa-miR-199b | CCCAGUGUUUAGACUAUCUGUUC | ||||
| miR-214 | 7025 | 7032 | 1A,m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
| hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG | ||||
| miR-218 | 450 | 456 | 1A | hsa-miR-218brain | UUGUGCUUGAUCUAACCAUGU |
| miR-22 | 7051 | 7058 | 1A,m8 | hsa-miR-22brain | AAGCUGCCAGUUGAAGAACUGU |
| miR-221/222 | 8360 | 8366 | 1A | hsa-miR-221brain | AGCUACAUUGUCUGCUGGGUUUC |
| hsa-miR-222brain | AGCUACAUCUGGCUACUGGGUCUC | ||||
| miR-30-3p | 7157 | 7164 | 1A,m8 | hsa-miR-30a-3p | CUUUCAGUCGGAUGUUUGCAGC |
| hsa-miR-30e-3p | CUUUCAGUCGGAUGUUUACAGC | ||||
| miR-320 | 8177 | 8183 | 1A | hsa-miR-320 | AAAAGCUGGGUUGAGAGGGCGAA |
| miR-330 | 3857 | 3863 | 1A | hsa-miR-330brain | GCAAAGCACACGGCCUGCAGAGA |
| miR-338 | 7619 | 7625 | 1A | hsa-miR-338brain | UCCAGCAUCAGUGAUUUUGUUGA |
| miR-374 | 404 | 410 | 1A | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-381 | 8007 | 8013 | 1A | hsa-miR-381 | UAUACAAGGGCAAGCUCUCUGU |
| miR-410 | 7112 | 7118 | 1A | hsa-miR-410 | AAUAUAACACAGAUGGCCUGU |
| miR-495 | 734 | 740 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
| miR-542-3p | 4125 | 4131 | m8 | hsa-miR-542-3p | UGUGACAGAUUGAUAACUGAAA |
| miR-543 | 8251 | 8257 | 1A | hsa-miR-543 | AAACAUUCGCGGUGCACUUCU |
| miR-9 | 6138 | 6144 | 1A | hsa-miR-9SZ | UCUUUGGUUAUCUAGCUGUAUGA |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.