Gene Page: SNX3
Summary ?
| GeneID | 8724 |
| Symbol | SNX3 |
| Synonyms | Grd19|MCOPS8|SDP3 |
| Description | sorting nexin 3 |
| Reference | MIM:605930|HGNC:HGNC:11174|Ensembl:ENSG00000112335|HPRD:09333|Vega:OTTHUMG00000015323 |
| Gene type | protein-coding |
| Map location | 6q21 |
| Pascal p-value | 0.279 |
| Fetal beta | -1.003 |
| DMG | 1 (# studies) |
| Support | G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| Expression | Meta-analysis of gene expression | P value: 1.513 | |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0044 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08417687 | 6 | 108582627 | SNX3 | 7.321E-4 | -5.052 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
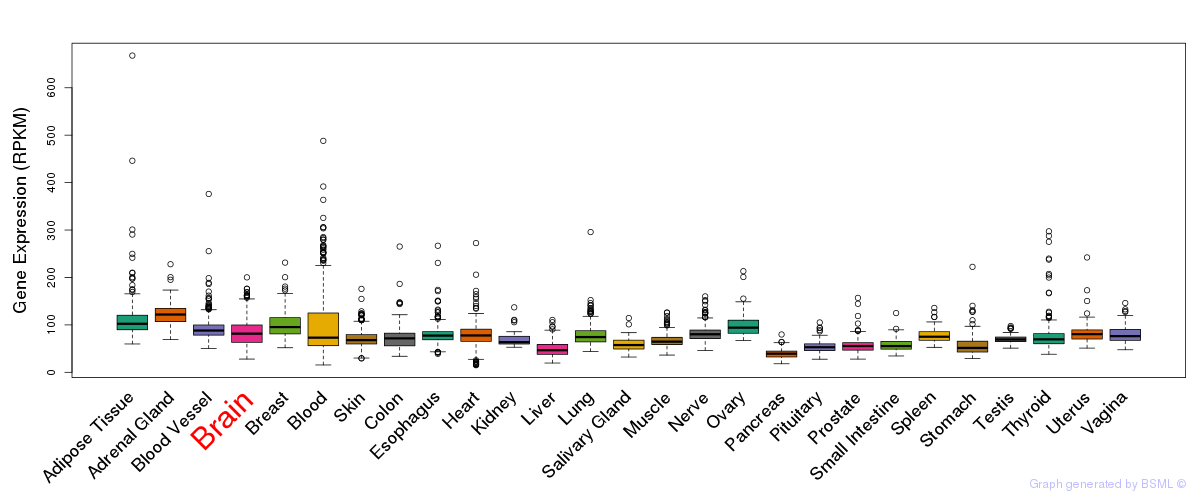
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GLUD1 | 0.90 | 0.88 |
| S1PR1 | 0.88 | 0.85 |
| GPC5 | 0.87 | 0.84 |
| GLUD2 | 0.87 | 0.85 |
| SLC1A3 | 0.87 | 0.89 |
| HEPH | 0.86 | 0.90 |
| PPAP2B | 0.86 | 0.88 |
| HTRA1 | 0.86 | 0.87 |
| ACSS3 | 0.85 | 0.90 |
| FAM167A | 0.85 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| DPF1 | -0.53 | -0.52 |
| MED19 | -0.52 | -0.56 |
| MARCH4 | -0.52 | -0.50 |
| MPP3 | -0.51 | -0.51 |
| ACTL6B | -0.51 | -0.53 |
| KIAA1543 | -0.51 | -0.48 |
| AC011491.1 | -0.51 | -0.56 |
| PAK7 | -0.50 | -0.47 |
| CABYR | -0.50 | -0.50 |
| ATXN7L1 | -0.50 | -0.47 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | IEA | - | |
| GO:0035091 | phosphoinositide binding | IEA | - | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007154 | cell communication | IEA | - | |
| GO:0006897 | endocytosis | TAS | 9819414 | |
| GO:0015031 | protein transport | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005737 | cytoplasm | IDA | 11279102 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| NAKAMURA TUMOR ZONE PERIPHERAL VS CENTRAL DN | 634 | 384 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| TIEN INTESTINE PROBIOTICS 24HR UP | 557 | 331 | All SZGR 2.0 genes in this pathway |
| KINSEY TARGETS OF EWSR1 FLII FUSION UP | 1278 | 748 | All SZGR 2.0 genes in this pathway |
| YANG BREAST CANCER ESR1 LASER DN | 50 | 38 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 6Q21 DELETION | 20 | 15 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC UP | 123 | 75 | All SZGR 2.0 genes in this pathway |
| DE YY1 TARGETS DN | 92 | 64 | All SZGR 2.0 genes in this pathway |
| BONOME OVARIAN CANCER SURVIVAL SUBOPTIMAL DEBULKING | 510 | 309 | All SZGR 2.0 genes in this pathway |
| TOOKER GEMCITABINE RESISTANCE DN | 122 | 84 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| AGUIRRE PANCREATIC CANCER COPY NUMBER DN | 238 | 145 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN PREDNISOLONE RESISTANCE B ALL UP | 22 | 16 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-142-5p | 532 | 538 | 1A | hsa-miR-142-5p | CAUAAAGUAGAAAGCACUAC |
| miR-539 | 364 | 370 | m8 | hsa-miR-539 | GGAGAAAUUAUCCUUGGUGUGU |
| miR-544 | 470 | 477 | 1A,m8 | hsa-miR-544 | AUUCUGCAUUUUUAGCAAGU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.