Gene Page: FADD
Summary ?
| GeneID | 8772 |
| Symbol | FADD |
| Synonyms | GIG3|MORT1 |
| Description | Fas associated via death domain |
| Reference | MIM:602457|HGNC:HGNC:3573|Ensembl:ENSG00000168040|HPRD:03909|Vega:OTTHUMG00000167264 |
| Gene type | protein-coding |
| Map location | 11q13.3 |
| Pascal p-value | 0.342 |
| Fetal beta | 0.445 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20821095 | 11 | 70049116 | FADD | 1.5E-11 | -0.016 | 2.86E-7 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
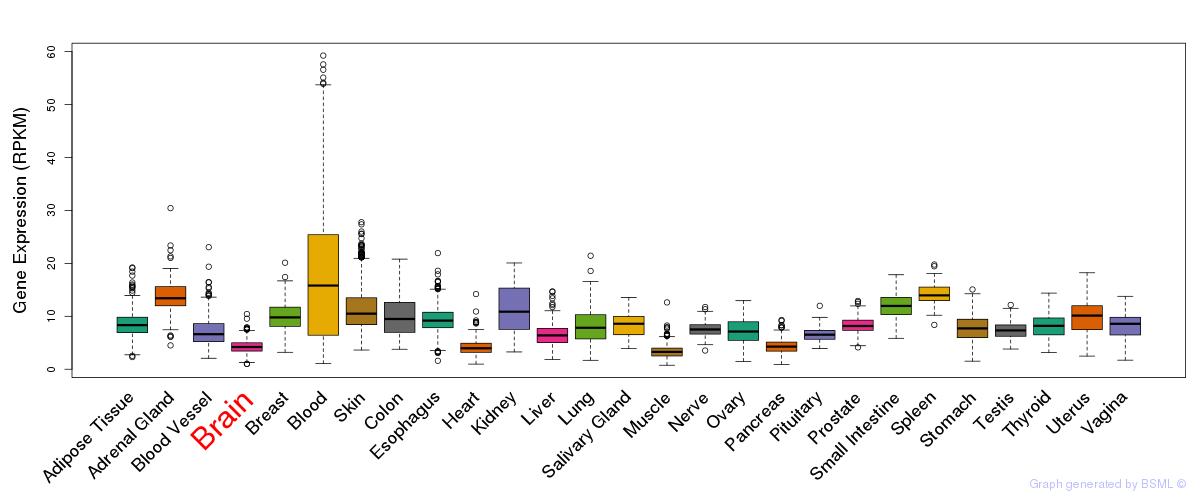
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABCA1 | ABC-1 | ABC1 | CERP | FLJ14958 | HDLDT1 | MGC164864 | MGC165011 | TGD | ATP-binding cassette, sub-family A (ABC1), member 1 | - | HPRD,BioGRID | 12235128 |
| ARHGDIA | GDIA1 | MGC117248 | RHOGDI | RHOGDI-1 | Rho GDP dissociation inhibitor (GDI) alpha | Co-purification | BioGRID | 15659383 |
| BID | FP497 | MGC15319 | MGC42355 | BH3 interacting domain death agonist | Co-purification | BioGRID | 15659383 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | - | HPRD | 9045686 |11717445|11717445 |
| CASP10 | ALPS2 | FLICE2 | MCH4 | caspase 10, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 11717445 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | CASP8 (caspase-8) interacts with FADD. | BIND | 15782135 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD,BioGRID | 12107169 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | - | HPRD | 11717445 |12107169|12107169 |
| CASP8 | ALPS2B | CAP4 | FLICE | FLJ17672 | MACH | MCH5 | MGC78473 | caspase 8, apoptosis-related cysteine peptidase | caspase-8 interacts with FADD. | BIND | 12220669 |
| CASP8AP2 | CED-4 | FLASH | FLJ11208 | KIAA1315 | RIP25 | caspase 8 associated protein 2 | - | HPRD,BioGRID | 10235259 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | MRIT-beta-1 interacts with FADD. This interaction was modelled on a demonstrated interaction between human MRIT-beta-1 and FADD from an unspecified species. | BIND | 9326610 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | - | HPRD | 9208847 |11965497 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | c-FLIP interacts with FADD. | BIND | 12220669 |
| CFLAR | CASH | CASP8AP1 | CLARP | Casper | FLAME | FLAME-1 | FLAME1 | FLIP | I-FLICE | MRIT | c-FLIP | c-FLIPL | c-FLIPR | c-FLIPS | CASP8 and FADD-like apoptosis regulator | Affinity Capture-Western Reconstituted Complex Two-hybrid | BioGRID | 9208847 |9228018 |9326610 |
| DAP3 | DAP-3 | DKFZp686G12159 | MGC126058 | MGC126059 | MRP-S29 | MRPS29 | bMRP-10 | death associated protein 3 | - | HPRD,BioGRID | 11376335 |11753396 |
| DAPK1 | DAPK | DKFZp781I035 | death-associated protein kinase 1 | - | HPRD,BioGRID | 12911633 |
| DEDD | CASP8IP1 | DEDD1 | DEFT | FLDED1 | KE05 | death effector domain containing | - | HPRD,BioGRID | 9774341 |
| EZR | CVIL | CVL | DKFZp762H157 | FLJ26216 | MGC1584 | VIL2 | ezrin | Co-purification | BioGRID | 15659383 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | Fas interacts with FADD. | BIND | 15383280 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | An unspecified isoform of Fas interacts with FADD. | BIND | 15665818 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD,BioGRID | 7538907 |8967952 |
| FAS | ALPS1A | APO-1 | APT1 | CD95 | FAS1 | FASTM | TNFRSF6 | Fas (TNF receptor superfamily, member 6) | - | HPRD | 7536190 |7538907 |8967952|7538907 |8967952 |
| FASLG | APT1LG1 | CD178 | CD95L | FASL | TNFSF6 | Fas ligand (TNF superfamily, member 6) | Affinity Capture-Western Co-purification | BioGRID | 12887920 |15659383 |
| HIPK3 | DYRK6 | FIST3 | PKY | YAK1 | homeodomain interacting protein kinase 3 | Affinity Capture-Western | BioGRID | 11034606 |
| IRAK1 | IRAK | pelle | interleukin-1 receptor-associated kinase 1 | - | HPRD,BioGRID | 11828002 |
| LRDD | DKFZp434D229 | MGC16925 | PIDD | leucine-rich repeats and death domain containing | - | HPRD,BioGRID | 10825539 |
| MAPK8 | JNK | JNK1 | JNK1A2 | JNK21B1/2 | PRKM8 | SAPK1 | mitogen-activated protein kinase 8 | Co-purification | BioGRID | 15659383 |
| MBD4 | MED1 | methyl-CpG binding domain protein 4 | - | HPRD,BioGRID | 12702765 |
| MOBKL3 | 2C4D | CGI-95 | MGC12264 | MOB1 | MOB3 | PREI3 | MOB1, Mps One Binder kinase activator-like 3 (yeast) | Two-hybrid | BioGRID | 16169070 |
| MSN | - | moesin | Co-purification | BioGRID | 15659383 |
| MYD88 | MYD88D | myeloid differentiation primary response gene (88) | - | HPRD,BioGRID | 11828002 |
| NACA | HSD48 | MGC117224 | NACA1 | nascent polypeptide-associated complex alpha subunit | - | HPRD,BioGRID | 12684039 |
| NOL3 | ARC | CARD2 | MYC | MYP | NOP | NOP30 | nucleolar protein 3 (apoptosis repressor with CARD domain) | ARC interacts with FADD. | BIND | 15383280 |
| NOL3 | ARC | CARD2 | MYC | MYP | NOP | NOP30 | nucleolar protein 3 (apoptosis repressor with CARD domain) | Phenotypic Suppression | BioGRID | 9560245 |
| PEA15 | HMAT1 | HUMMAT1H | MAT1 | MAT1H | PEA-15 | PED | phosphoprotein enriched in astrocytes 15 | - | HPRD,BioGRID | 10442631 |10493725 |
| PRKCZ | PKC-ZETA | PKC2 | protein kinase C, zeta | - | HPRD,BioGRID | 11739185 |
| RHOA | ARH12 | ARHA | RHO12 | RHOH12 | ras homolog gene family, member A | Co-purification | BioGRID | 15659383 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | Two-hybrid | BioGRID | 11854271 |
| RIPK1 | FLJ39204 | RIP | RIP1 | receptor (TNFRSF)-interacting serine-threonine kinase 1 | - | HPRD | 11002422 |
| RYBP | AAP1 | DEDAF | YEAF1 | RING1 and YY1 binding protein | Affinity Capture-Western | BioGRID | 11395500 |
| SUMO1 | DAP-1 | GMP1 | OFC10 | PIC1 | SENP2 | SMT3 | SMT3C | SMT3H3 | SUMO-1 | UBL1 | SMT3 suppressor of mif two 3 homolog 1 (S. cerevisiae) | Two-hybrid | BioGRID | 8906799 |
| TNFRSF10A | APO2 | CD261 | DR4 | MGC9365 | TRAILR-1 | TRAILR1 | tumor necrosis factor receptor superfamily, member 10a | - | HPRD,BioGRID | 9430227 |11376335 |
| TNFRSF10B | CD262 | DR5 | KILLER | KILLER/DR5 | TRAIL-R2 | TRAILR2 | TRICK2 | TRICK2A | TRICK2B | TRICKB | ZTNFR9 | tumor necrosis factor receptor superfamily, member 10b | - | HPRD,BioGRID | 9430227 |
| TNFRSF1A | CD120a | FPF | MGC19588 | TBP1 | TNF-R | TNF-R-I | TNF-R55 | TNFAR | TNFR1 | TNFR55 | TNFR60 | p55 | p55-R | p60 | tumor necrosis factor receptor superfamily, member 1A | Affinity Capture-Western Co-purification | BioGRID | 8565075 |15659383 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | TRADD interacts with FADD. This interaction was modeled on a demonstrated interaction between TRADD and FADD, both from an unspecified species | BIND | 12796506 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | Affinity Capture-Western Two-hybrid | BioGRID | 8565075 |12911633 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | FADD interacts with TRADD. | BIND | 11112409 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | TRADD interacts with FADD. | BIND | 15761471 |
| TRADD | Hs.89862 | MGC11078 | TNFRSF1A-associated via death domain | - | HPRD | 8565075 |10911999 |
| XPO5 | FLJ14239 | FLJ32057 | FLJ45606 | KIAA1291 | exportin 5 | - | HPRD | 12702765 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG APOPTOSIS | 88 | 62 | All SZGR 2.0 genes in this pathway |
| KEGG TOLL LIKE RECEPTOR SIGNALING PATHWAY | 102 | 88 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| KEGG ALZHEIMERS DISEASE | 169 | 110 | All SZGR 2.0 genes in this pathway |
| KEGG PATHWAYS IN CANCER | 328 | 259 | All SZGR 2.0 genes in this pathway |
| BIOCARTA RELA PATHWAY | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CERAMIDE PATHWAY | 22 | 16 | All SZGR 2.0 genes in this pathway |
| BIOCARTA FAS PATHWAY | 30 | 22 | All SZGR 2.0 genes in this pathway |
| BIOCARTA HIVNEF PATHWAY | 58 | 43 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DEATH PATHWAY | 33 | 24 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NFKB PATHWAY | 23 | 19 | All SZGR 2.0 genes in this pathway |
| BIOCARTA SODD PATHWAY | 10 | 7 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TNFR1 PATHWAY | 29 | 21 | All SZGR 2.0 genes in this pathway |
| ST TUMOR NECROSIS FACTOR PATHWAY | 29 | 20 | All SZGR 2.0 genes in this pathway |
| ST FAS SIGNALING PATHWAY | 65 | 54 | All SZGR 2.0 genes in this pathway |
| PID TRAIL PATHWAY | 28 | 20 | All SZGR 2.0 genes in this pathway |
| PID FAS PATHWAY | 38 | 29 | All SZGR 2.0 genes in this pathway |
| PID TNF PATHWAY | 46 | 33 | All SZGR 2.0 genes in this pathway |
| PID CERAMIDE PATHWAY | 48 | 37 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME APOPTOSIS | 148 | 94 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS UP | 579 | 346 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 UP | 380 | 236 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| LAU APOPTOSIS CDKN2A UP | 55 | 40 | All SZGR 2.0 genes in this pathway |
| NADERI BREAST CANCER PROGNOSIS UP | 50 | 26 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| JAERVINEN AMPLIFIED IN LARYNGEAL CANCER | 40 | 24 | All SZGR 2.0 genes in this pathway |
| LOCKWOOD AMPLIFIED IN LUNG CANCER | 214 | 139 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| HOFMANN CELL LYMPHOMA DN | 39 | 29 | All SZGR 2.0 genes in this pathway |
| THEILGAARD NEUTROPHIL AT SKIN WOUND DN | 225 | 163 | All SZGR 2.0 genes in this pathway |
| DAZARD RESPONSE TO UV SCC DN | 123 | 86 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C2 | 18 | 15 | All SZGR 2.0 genes in this pathway |
| GENTILE UV RESPONSE CLUSTER D5 | 39 | 26 | All SZGR 2.0 genes in this pathway |
| GENTILE UV HIGH DOSE DN | 312 | 203 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 6HR DN | 160 | 101 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY E2F4 UNSTIMULATED | 728 | 415 | All SZGR 2.0 genes in this pathway |
| LIN MELANOMA COPY NUMBER UP | 73 | 53 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM BIPOLAR DISORDER OLIGODENDROCYTE DENSITY CORR UP | 682 | 440 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| LIM MAMMARY STEM CELL DN | 428 | 246 | All SZGR 2.0 genes in this pathway |