Gene Page: RGS11
Summary ?
| GeneID | 8786 |
| Symbol | RGS11 |
| Synonyms | RS11 |
| Description | regulator of G-protein signaling 11 |
| Reference | MIM:603895|HGNC:HGNC:9993|Ensembl:ENSG00000076344|HPRD:04872|Vega:OTTHUMG00000064893 |
| Gene type | protein-coding |
| Map location | 16p13.3 |
| Pascal p-value | 0.219 |
| Sherlock p-value | 0.514 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2502827 | chr1 | 176044216 | RGS11 | 8786 | 0.15 | trans | ||
| rs16829545 | chr2 | 151977407 | RGS11 | 8786 | 4.972E-7 | trans | ||
| rs3845734 | chr2 | 171125572 | RGS11 | 8786 | 0 | trans | ||
| rs7584986 | chr2 | 184111432 | RGS11 | 8786 | 0.01 | trans | ||
| rs17762315 | chr5 | 76807576 | RGS11 | 8786 | 0.06 | trans | ||
| rs16878286 | chr8 | 31646175 | RGS11 | 8786 | 0.11 | trans | ||
| rs16878317 | chr8 | 31657853 | RGS11 | 8786 | 0.05 | trans | ||
| rs16955618 | chr15 | 29937543 | RGS11 | 8786 | 6.57E-12 | trans |
Section II. Transcriptome annotation
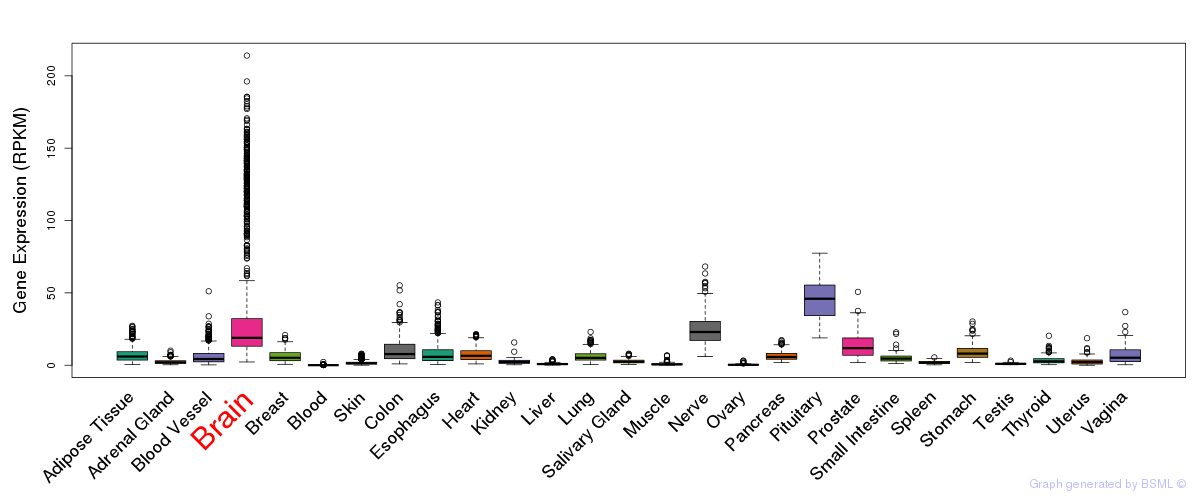
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TOM1L2 | 0.83 | 0.86 |
| SYNM | 0.82 | 0.80 |
| FAM120A | 0.82 | 0.84 |
| MAP3K5 | 0.81 | 0.82 |
| SIRPA | 0.80 | 0.85 |
| BCL2L2 | 0.80 | 0.79 |
| KIAA1671 | 0.80 | 0.84 |
| ASPH | 0.80 | 0.83 |
| UTRN | 0.79 | 0.83 |
| SLC6A1 | 0.79 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| RPL35 | -0.56 | -0.65 |
| RPL27 | -0.56 | -0.64 |
| RPL31 | -0.55 | -0.63 |
| RPL36 | -0.55 | -0.65 |
| RPS18 | -0.55 | -0.63 |
| RPL24 | -0.54 | -0.62 |
| RPS23 | -0.54 | -0.63 |
| RPL18 | -0.54 | -0.64 |
| RPS3AP47 | -0.54 | -0.58 |
| RPL32 | -0.53 | -0.61 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME SIGNALING BY GPCR | 920 | 449 | All SZGR 2.0 genes in this pathway |
| REACTOME GPCR DOWNSTREAM SIGNALING | 805 | 368 | All SZGR 2.0 genes in this pathway |
| REACTOME G ALPHA I SIGNALLING EVENTS | 195 | 114 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LINDGREN BLADDER CANCER CLUSTER 2B | 392 | 251 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO BEXAROTENE UP | 34 | 17 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 16P13 AMPLICON | 120 | 49 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| GAVIN FOXP3 TARGETS CLUSTER P2 | 79 | 55 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER BASAL DN | 701 | 446 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 2ND EGF PULSE ONLY | 1725 | 838 | All SZGR 2.0 genes in this pathway |