Gene Page: SAP30
Summary ?
| GeneID | 8819 |
| Symbol | SAP30 |
| Synonyms | - |
| Description | Sin3A associated protein 30kDa |
| Reference | MIM:603378|HGNC:HGNC:10532|Ensembl:ENSG00000164105|HPRD:04540|Vega:OTTHUMG00000160798 |
| Gene type | protein-coding |
| Map location | 4q34.1 |
| Pascal p-value | 0.234 |
| Sherlock p-value | 0.649 |
| DEG p-value | DEG:Maycox_2009:CC_BA10_fold_change=1.10:CC_BA10_disease_P=0.0370:HBB_BA9_fold_change=1.21:HBB_BA9_disease_P=0.0274 |
| Fetal beta | 0.742 |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DEG:Maycox_2009 | Microarray to determine the expression of over 30000 mRNA transcripts in post-mortem tissue | We included 51 genes whose expression changes are common between two schizophrenia cohorts. |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1561519 | chr2 | 35823020 | SAP30 | 8819 | 0.11 | trans | ||
| rs7591339 | chr2 | 35825597 | SAP30 | 8819 | 0.11 | trans | ||
| rs7570146 | chr2 | 35844751 | SAP30 | 8819 | 0.11 | trans | ||
| rs1439691 | chr2 | 35851281 | SAP30 | 8819 | 0.11 | trans |
Section II. Transcriptome annotation
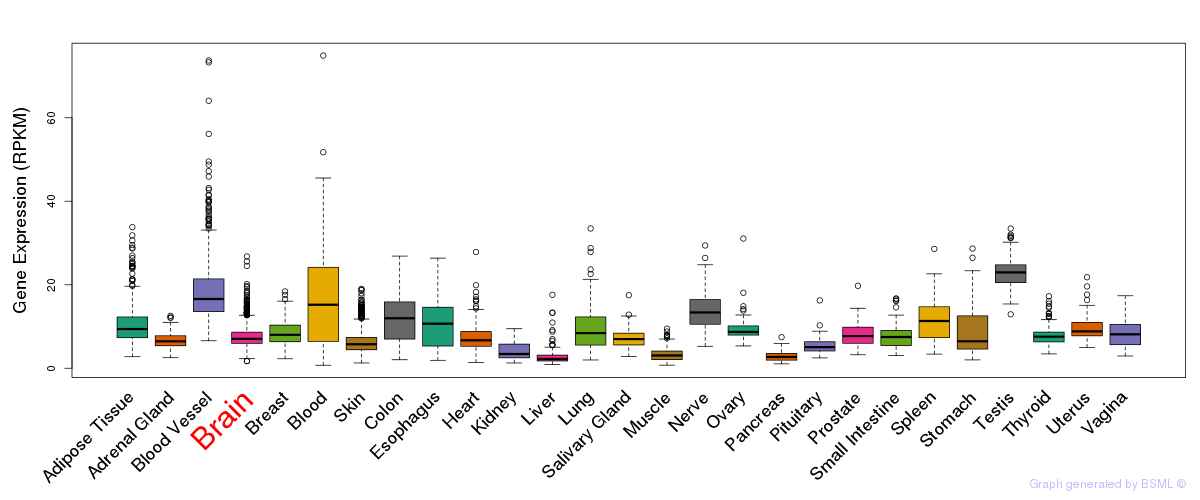
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| USP33 | 0.94 | 0.95 |
| ARCN1 | 0.93 | 0.93 |
| ATAD1 | 0.93 | 0.91 |
| SHOC2 | 0.93 | 0.93 |
| CAB39 | 0.92 | 0.93 |
| LRPPRC | 0.92 | 0.93 |
| STAU2 | 0.92 | 0.92 |
| SLC30A9 | 0.92 | 0.92 |
| ADSS | 0.91 | 0.91 |
| ARMC8 | 0.91 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.73 | -0.74 |
| AF347015.21 | -0.72 | -0.72 |
| FXYD1 | -0.72 | -0.74 |
| AF347015.31 | -0.72 | -0.73 |
| AF347015.8 | -0.71 | -0.73 |
| HIGD1B | -0.70 | -0.72 |
| EIF4EBP3 | -0.69 | -0.74 |
| IFI27 | -0.69 | -0.72 |
| MT-CYB | -0.69 | -0.70 |
| AF347015.2 | -0.69 | -0.70 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CIR | - | CBF1 interacting corepressor | - | HPRD,BioGRID | 9874765 |
| CIR | - | CBF1 interacting corepressor | CIR interacts with SAP30 | BIND | 11000236 |
| HBP1 | FLJ16340 | HMG-box transcription factor 1 | - | HPRD | 15235594 |
| HDAC1 | DKFZp686H12203 | GON-10 | HD1 | RPD3 | RPD3L1 | histone deacetylase 1 | - | HPRD,BioGRID | 9651585 |
| HDAC2 | RPD3 | YAF1 | histone deacetylase 2 | Affinity Capture-MS Affinity Capture-Western Co-purification | BioGRID | 9651585 |10444591 |11784859 |
| HIST4H4 | H4/p | MGC24116 | histone cluster 4, H4 | - | HPRD | 9651585 |
| ING1 | p24ING1c | p33 | p33ING1 | p33ING1b | p47 | p47ING1a | inhibitor of growth family, member 1 | - | HPRD,BioGRID | 11784859 |
| MTA2 | DKFZp686F2281 | MTA1L1 | PID | metastasis associated 1 family, member 2 | Co-purification | BioGRID | 10444591 |
| MXD1 | MAD | MAD1 | MGC104659 | MAX dimerization protein 1 | Reconstituted Complex | BioGRID | 11101889 |
| NCOR1 | KIAA1047 | MGC104216 | N-CoR | TRAC1 | hCIT529I10 | hN-CoR | nuclear receptor co-repressor 1 | - | HPRD,BioGRID | 9702189 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | - | HPRD | 9702189 |
| NCOR2 | CTG26 | SMRT | SMRTE | SMRTE-tau | TNRC14 | TRAC-1 | TRAC1 | nuclear receptor co-repressor 2 | Co-purification | BioGRID | 11013263 |
| PHF12 | FLJ34122 | KIAA1523 | MGC131914 | PF1 | PHD finger protein 12 | - | HPRD,BioGRID | 11390640 |
| RBBP4 | NURF55 | RBAP48 | retinoblastoma binding protein 4 | - | HPRD,BioGRID | 9651585 |
| RBBP7 | MGC138867 | MGC138868 | RbAp46 | retinoblastoma binding protein 7 | Affinity Capture-MS Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 9651585 |10444591 |11784859 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | - | HPRD | 11013263 |
| SIN3A | DKFZp434K2235 | FLJ90319 | KIAA0700 | SIN3 homolog A, transcription regulator (yeast) | Affinity Capture-MS Affinity Capture-Western Co-purification Reconstituted Complex | BioGRID | 9651585 |9702189 |10444591 |11390640 |11784859 |12724404 |
| SIN3B | KIAA0700 | SIN3 homolog B, transcription regulator (yeast) | Co-purification | BioGRID | 10444591 |
| YY1 | DELTA | INO80S | NF-E1 | UCRBP | YIN-YANG-1 | YY1 transcription factor | - | HPRD,BioGRID | 12788099 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| PID SMAD2 3NUCLEAR PATHWAY | 82 | 63 | All SZGR 2.0 genes in this pathway |
| PID HDAC CLASSI PATHWAY | 66 | 50 | All SZGR 2.0 genes in this pathway |
| PID TELOMERASE PATHWAY | 68 | 48 | All SZGR 2.0 genes in this pathway |
| PID HEDGEHOG GLI PATHWAY | 48 | 35 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| SENGUPTA NASOPHARYNGEAL CARCINOMA WITH LMP1 UP | 408 | 247 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES UP | 82 | 51 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA UP | 177 | 110 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| LIU CMYB TARGETS UP | 165 | 106 | All SZGR 2.0 genes in this pathway |
| LIU VMYB TARGETS UP | 127 | 78 | All SZGR 2.0 genes in this pathway |
| BORCZUK MALIGNANT MESOTHELIOMA UP | 305 | 185 | All SZGR 2.0 genes in this pathway |
| OSMAN BLADDER CANCER UP | 404 | 246 | All SZGR 2.0 genes in this pathway |
| DITTMER PTHLH TARGETS DN | 73 | 51 | All SZGR 2.0 genes in this pathway |
| MARKEY RB1 ACUTE LOF UP | 215 | 137 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA UP | 536 | 340 | All SZGR 2.0 genes in this pathway |
| MATTIOLI MGUS VS PCL | 116 | 62 | All SZGR 2.0 genes in this pathway |
| MARTORIATI MDM4 TARGETS FETAL LIVER UP | 227 | 137 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 6HR DN | 514 | 330 | All SZGR 2.0 genes in this pathway |
| BENPORATH SOX2 TARGETS | 734 | 436 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| BENPORATH CYCLING GENES | 648 | 385 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES CORE NINE CORRELATED | 100 | 68 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SASAKI ADULT T CELL LEUKEMIA | 176 | 122 | All SZGR 2.0 genes in this pathway |
| NEMETH INFLAMMATORY RESPONSE LPS UP | 88 | 64 | All SZGR 2.0 genes in this pathway |
| MOREAUX MULTIPLE MYELOMA BY TACI UP | 412 | 249 | All SZGR 2.0 genes in this pathway |
| KIM HYPOXIA | 25 | 21 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS LATE PROGENITOR | 544 | 307 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP | 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| LEONARD HYPOXIA | 47 | 35 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| YAMAZAKI TCEB3 TARGETS DN | 215 | 132 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| ZHANG BREAST CANCER PROGENITORS UP | 425 | 253 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A DN | 159 | 105 | All SZGR 2.0 genes in this pathway |
| HOFFMANN PRE BI TO LARGE PRE BII LYMPHOCYTE UP | 36 | 24 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH T 8 21 TRANSLOCATION | 368 | 247 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| WHITFIELD CELL CYCLE G2 | 182 | 102 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| SUBTIL PROGESTIN TARGETS | 36 | 25 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE MIDDLE | 98 | 56 | All SZGR 2.0 genes in this pathway |
| MIYAGAWA TARGETS OF EWSR1 ETS FUSIONS UP | 259 | 159 | All SZGR 2.0 genes in this pathway |
| PEDERSEN METASTASIS BY ERBB2 ISOFORM 7 | 403 | 240 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG BOUND 8D | 658 | 397 | All SZGR 2.0 genes in this pathway |
| KRIEG HYPOXIA NOT VIA KDM3A | 770 | 480 | All SZGR 2.0 genes in this pathway |
| FOSTER KDM1A TARGETS DN | 211 | 119 | All SZGR 2.0 genes in this pathway |
| ALTEMEIER RESPONSE TO LPS WITH MECHANICAL VENTILATION | 128 | 81 | All SZGR 2.0 genes in this pathway |
| ZHOU CELL CYCLE GENES IN IR RESPONSE 6HR | 85 | 49 | All SZGR 2.0 genes in this pathway |
| ZWANG TRANSIENTLY UP BY 1ST EGF PULSE ONLY | 1839 | 928 | All SZGR 2.0 genes in this pathway |