Gene Page: RSPH1
Summary ?
| GeneID | 89765 |
| Symbol | RSPH1 |
| Synonyms | CT79|RSP44|RSPH10A|TSA2|TSGA2 |
| Description | radial spoke head 1 homolog (Chlamydomonas) |
| Reference | MIM:609314|HGNC:HGNC:12371|Ensembl:ENSG00000160188|HPRD:11651|Vega:OTTHUMG00000086804 |
| Gene type | protein-coding |
| Map location | 21q22.3 |
| Pascal p-value | 0.187 |
| Sherlock p-value | 0.027 |
| eGene | Caudate basal ganglia Cortex Frontal Cortex BA9 Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs12122035 | chr1 | 50726051 | RSPH1 | 89765 | 0.06 | trans | ||
| rs6713473 | chr2 | 11284414 | RSPH1 | 89765 | 0.2 | trans | ||
| rs7578306 | chr2 | 171082977 | RSPH1 | 89765 | 0.02 | trans | ||
| rs6799944 | chr3 | 122905753 | RSPH1 | 89765 | 0.14 | trans | ||
| rs2268637 | chr6 | 39050917 | RSPH1 | 89765 | 0.05 | trans | ||
| rs11966129 | chr6 | 138693996 | RSPH1 | 89765 | 0.17 | trans | ||
| rs1341140 | chr13 | 79534499 | RSPH1 | 89765 | 0.14 | trans | ||
| rs12585805 | chr13 | 79546978 | RSPH1 | 89765 | 0.14 | trans | ||
| rs9976598 | 21 | 43901777 | RSPH1 | ENSG00000160188.5 | 3.313E-7 | 0.01 | 11412 | gtex_brain_putamen_basal |
| rs200939470 | 21 | 43903570 | RSPH1 | ENSG00000160188.5 | 4.953E-8 | 0.01 | 9619 | gtex_brain_putamen_basal |
| rs66906364 | 21 | 43903571 | RSPH1 | ENSG00000160188.5 | 4.942E-8 | 0.01 | 9618 | gtex_brain_putamen_basal |
| rs2839536 | 21 | 43905887 | RSPH1 | ENSG00000160188.5 | 4.461E-8 | 0.01 | 7302 | gtex_brain_putamen_basal |
| rs2839537 | 21 | 43906351 | RSPH1 | ENSG00000160188.5 | 4.461E-8 | 0.01 | 6838 | gtex_brain_putamen_basal |
| rs4920113 | 21 | 43907252 | RSPH1 | ENSG00000160188.5 | 4.461E-8 | 0.01 | 5937 | gtex_brain_putamen_basal |
| rs9983237 | 21 | 43909302 | RSPH1 | ENSG00000160188.5 | 4.461E-8 | 0.01 | 3887 | gtex_brain_putamen_basal |
| rs9979100 | 21 | 43913370 | RSPH1 | ENSG00000160188.5 | 1.698E-7 | 0.01 | -181 | gtex_brain_putamen_basal |
| rs1573413 | 21 | 43916339 | RSPH1 | ENSG00000160188.5 | 7.753E-8 | 0.01 | -3150 | gtex_brain_putamen_basal |
| rs9984443 | 21 | 43929514 | RSPH1 | ENSG00000160188.5 | 1.843E-7 | 0.01 | -16325 | gtex_brain_putamen_basal |
| rs8126600 | 21 | 43929852 | RSPH1 | ENSG00000160188.5 | 1.841E-7 | 0.01 | -16663 | gtex_brain_putamen_basal |
| rs4919990 | 21 | 43938673 | RSPH1 | ENSG00000160188.5 | 4.48E-7 | 0.01 | -25484 | gtex_brain_putamen_basal |
| rs3788020 | 21 | 43947353 | RSPH1 | ENSG00000160188.5 | 6.05E-8 | 0.01 | -34164 | gtex_brain_putamen_basal |
| rs10887974 | 21 | 43949715 | RSPH1 | ENSG00000160188.5 | 1.152E-7 | 0.01 | -36526 | gtex_brain_putamen_basal |
| rs228049 | 21 | 43952437 | RSPH1 | ENSG00000160188.5 | 3.79E-7 | 0.01 | -39248 | gtex_brain_putamen_basal |
| rs228081 | 21 | 43966971 | RSPH1 | ENSG00000160188.5 | 2.361E-7 | 0.01 | -53782 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
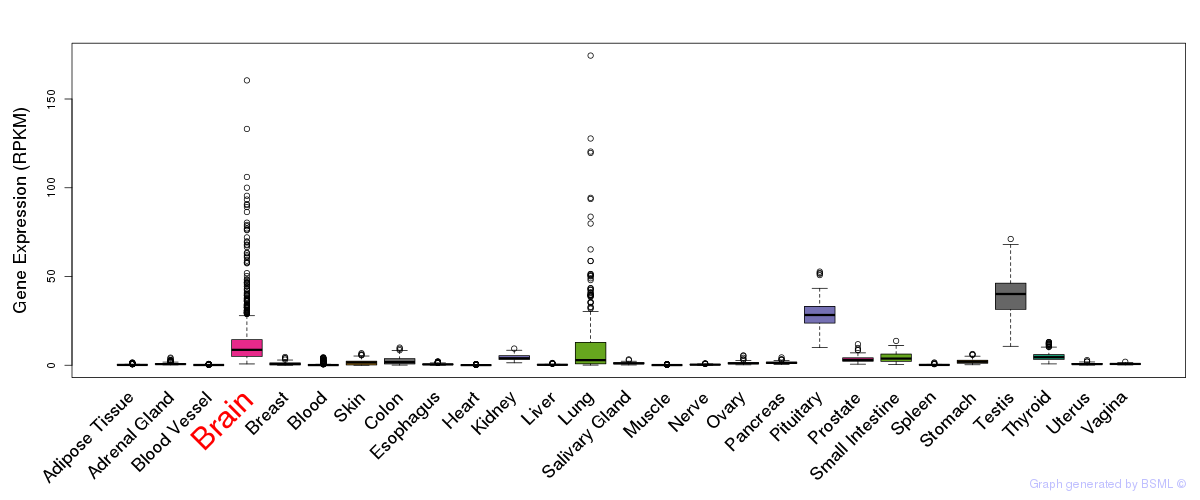
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| SENGUPTA NASOPHARYNGEAL CARCINOMA DN | 349 | 157 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS BASAL UP | 380 | 215 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL UP | 450 | 256 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA UP | 1821 | 933 | All SZGR 2.0 genes in this pathway |
| LIEN BREAST CARCINOMA METAPLASTIC VS DUCTAL DN | 114 | 58 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 2 | 473 | 224 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA D CLUSTER DN | 40 | 26 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D DN | 252 | 155 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA PCA3 DN | 69 | 38 | All SZGR 2.0 genes in this pathway |