Gene Page: MCFD2
Summary ?
| GeneID | 90411 |
| Symbol | MCFD2 |
| Synonyms | F5F8D|F5F8D2|LMAN1IP|SDNSF |
| Description | multiple coagulation factor deficiency 2 |
| Reference | MIM:607788|HGNC:HGNC:18451|Ensembl:ENSG00000180398|HPRD:09693|Vega:OTTHUMG00000153100 |
| Gene type | protein-coding |
| Map location | 2p21 |
| Pascal p-value | 0.463 |
| DMG | 1 (# studies) |
| eGene | Anterior cingulate cortex BA24 Caudate basal ganglia Cerebellar Hemisphere Cerebellum Cortex Frontal Cortex BA9 Hippocampus Nucleus accumbens basal ganglia Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg14126210 | 2 | 47142867 | MCFD2 | -0.018 | 0.27 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11890177 | 2 | 47133055 | MCFD2 | ENSG00000180398.7 | 2.119E-11 | 0 | 10196 | gtex_brain_ba24 |
| rs35433380 | 2 | 47136950 | MCFD2 | ENSG00000180398.7 | 1.17E-10 | 0 | 6301 | gtex_brain_ba24 |
| rs62136827 | 2 | 47137167 | MCFD2 | ENSG00000180398.7 | 9.521E-7 | 0 | 6084 | gtex_brain_ba24 |
| rs34581142 | 2 | 47140612 | MCFD2 | ENSG00000180398.7 | 3.181E-8 | 0 | 2639 | gtex_brain_ba24 |
| rs199965792 | 2 | 47142958 | MCFD2 | ENSG00000180398.7 | 1.321E-6 | 0 | 293 | gtex_brain_ba24 |
| rs6544939 | 2 | 47143230 | MCFD2 | ENSG00000180398.7 | 7.902E-8 | 0 | 21 | gtex_brain_ba24 |
| rs6733548 | 2 | 47143929 | MCFD2 | ENSG00000180398.7 | 5.443E-7 | 0 | -678 | gtex_brain_ba24 |
| rs6716381 | 2 | 47148163 | MCFD2 | ENSG00000180398.7 | 4.497E-12 | 0 | -4912 | gtex_brain_ba24 |
| rs397872646 | 2 | 47149036 | MCFD2 | ENSG00000180398.7 | 8.709E-8 | 0 | -5785 | gtex_brain_ba24 |
| rs397871037 | 2 | 47149693 | MCFD2 | ENSG00000180398.7 | 4.497E-12 | 0 | -6442 | gtex_brain_ba24 |
| rs70940647 | 2 | 47150500 | MCFD2 | ENSG00000180398.7 | 1.034E-7 | 0 | -7249 | gtex_brain_ba24 |
| rs7601491 | 2 | 47150626 | MCFD2 | ENSG00000180398.7 | 2.119E-11 | 0 | -7375 | gtex_brain_ba24 |
| rs13384023 | 2 | 47153596 | MCFD2 | ENSG00000180398.7 | 1.075E-6 | 0 | -10345 | gtex_brain_ba24 |
| rs55688290 | 2 | 47154113 | MCFD2 | ENSG00000180398.7 | 1.075E-6 | 0 | -10862 | gtex_brain_ba24 |
| rs56176930 | 2 | 47155868 | MCFD2 | ENSG00000180398.7 | 1.169E-8 | 0 | -12617 | gtex_brain_ba24 |
| rs7604228 | 2 | 47156684 | MCFD2 | ENSG00000180398.7 | 1.075E-6 | 0 | -13433 | gtex_brain_ba24 |
| rs13398851 | 2 | 47160016 | MCFD2 | ENSG00000180398.7 | 3.275E-7 | 0 | -16765 | gtex_brain_ba24 |
| rs13392448 | 2 | 47166410 | MCFD2 | ENSG00000180398.7 | 8.709E-8 | 0 | -23159 | gtex_brain_ba24 |
| rs6755303 | 2 | 47169003 | MCFD2 | ENSG00000180398.7 | 8.709E-8 | 0 | -25752 | gtex_brain_ba24 |
| rs7574514 | 2 | 47125914 | MCFD2 | ENSG00000180398.7 | 1.333E-6 | 0 | 17337 | gtex_brain_putamen_basal |
| rs10199965 | 2 | 47128603 | MCFD2 | ENSG00000180398.7 | 1.288E-6 | 0 | 14648 | gtex_brain_putamen_basal |
| rs28516550 | 2 | 47131909 | MCFD2 | ENSG00000180398.7 | 1.611E-6 | 0 | 11342 | gtex_brain_putamen_basal |
| rs11890177 | 2 | 47133055 | MCFD2 | ENSG00000180398.7 | 1.03E-11 | 0 | 10196 | gtex_brain_putamen_basal |
| rs62136823 | 2 | 47134390 | MCFD2 | ENSG00000180398.7 | 1.611E-6 | 0 | 8861 | gtex_brain_putamen_basal |
| rs2289928 | 2 | 47134904 | MCFD2 | ENSG00000180398.7 | 1.611E-6 | 0 | 8347 | gtex_brain_putamen_basal |
| rs2289927 | 2 | 47135246 | MCFD2 | ENSG00000180398.7 | 1.611E-6 | 0 | 8005 | gtex_brain_putamen_basal |
| rs7565420 | 2 | 47135769 | MCFD2 | ENSG00000180398.7 | 1.557E-6 | 0 | 7482 | gtex_brain_putamen_basal |
| rs62136825 | 2 | 47136617 | MCFD2 | ENSG00000180398.7 | 1.479E-6 | 0 | 6634 | gtex_brain_putamen_basal |
| rs79572152 | 2 | 47136638 | MCFD2 | ENSG00000180398.7 | 1.477E-6 | 0 | 6613 | gtex_brain_putamen_basal |
| rs35433380 | 2 | 47136950 | MCFD2 | ENSG00000180398.7 | 1.052E-10 | 0 | 6301 | gtex_brain_putamen_basal |
| rs62136827 | 2 | 47137167 | MCFD2 | ENSG00000180398.7 | 1.446E-6 | 0 | 6084 | gtex_brain_putamen_basal |
| rs11894319 | 2 | 47137267 | MCFD2 | ENSG00000180398.7 | 1.428E-6 | 0 | 5984 | gtex_brain_putamen_basal |
| rs6722440 | 2 | 47137842 | MCFD2 | ENSG00000180398.7 | 1.374E-6 | 0 | 5409 | gtex_brain_putamen_basal |
| rs10168894 | 2 | 47139018 | MCFD2 | ENSG00000180398.7 | 1.581E-6 | 0 | 4233 | gtex_brain_putamen_basal |
| rs6758512 | 2 | 47139183 | MCFD2 | ENSG00000180398.7 | 1.284E-6 | 0 | 4068 | gtex_brain_putamen_basal |
| rs10174894 | 2 | 47140152 | MCFD2 | ENSG00000180398.7 | 1.243E-6 | 0 | 3099 | gtex_brain_putamen_basal |
| rs34581142 | 2 | 47140612 | MCFD2 | ENSG00000180398.7 | 1.267E-8 | 0 | 2639 | gtex_brain_putamen_basal |
| rs6706031 | 2 | 47140978 | MCFD2 | ENSG00000180398.7 | 4.38E-7 | 0 | 2273 | gtex_brain_putamen_basal |
| rs6544939 | 2 | 47143230 | MCFD2 | ENSG00000180398.7 | 6.259E-9 | 0 | 21 | gtex_brain_putamen_basal |
| rs62140550 | 2 | 47143989 | MCFD2 | ENSG00000180398.7 | 1.346E-6 | 0 | -738 | gtex_brain_putamen_basal |
| rs13388612 | 2 | 47144203 | MCFD2 | ENSG00000180398.7 | 1.365E-6 | 0 | -952 | gtex_brain_putamen_basal |
| rs6758093 | 2 | 47144862 | MCFD2 | ENSG00000180398.7 | 1.411E-6 | 0 | -1611 | gtex_brain_putamen_basal |
| rs6729604 | 2 | 47145002 | MCFD2 | ENSG00000180398.7 | 1.411E-6 | 0 | -1751 | gtex_brain_putamen_basal |
| rs6716381 | 2 | 47148163 | MCFD2 | ENSG00000180398.7 | 1.03E-11 | 0 | -4912 | gtex_brain_putamen_basal |
| rs397872646 | 2 | 47149036 | MCFD2 | ENSG00000180398.7 | 3.894E-7 | 0 | -5785 | gtex_brain_putamen_basal |
| rs397871037 | 2 | 47149693 | MCFD2 | ENSG00000180398.7 | 1.03E-11 | 0 | -6442 | gtex_brain_putamen_basal |
| rs70940647 | 2 | 47150500 | MCFD2 | ENSG00000180398.7 | 1.496E-8 | 0 | -7249 | gtex_brain_putamen_basal |
| rs7601491 | 2 | 47150626 | MCFD2 | ENSG00000180398.7 | 1.03E-11 | 0 | -7375 | gtex_brain_putamen_basal |
| rs13398851 | 2 | 47160016 | MCFD2 | ENSG00000180398.7 | 7.576E-8 | 0 | -16765 | gtex_brain_putamen_basal |
| rs13392448 | 2 | 47166410 | MCFD2 | ENSG00000180398.7 | 1.945E-8 | 0 | -23159 | gtex_brain_putamen_basal |
| rs6755303 | 2 | 47169003 | MCFD2 | ENSG00000180398.7 | 1.945E-8 | 0 | -25752 | gtex_brain_putamen_basal |
| rs9973504 | 2 | 47171955 | MCFD2 | ENSG00000180398.7 | 1.196E-6 | 0 | -28704 | gtex_brain_putamen_basal |
| rs5830929 | 2 | 47172873 | MCFD2 | ENSG00000180398.7 | 4.222E-9 | 0 | -29622 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
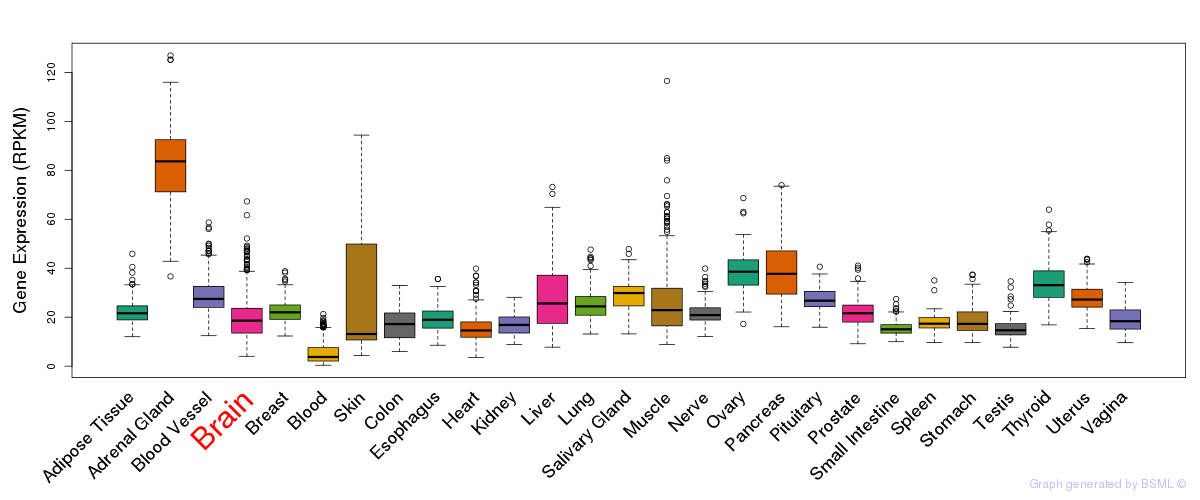
General gene expression (GTEx)
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| TLN1 | 0.81 | 0.84 |
| RXRA | 0.80 | 0.83 |
| FYCO1 | 0.79 | 0.84 |
| CDH20 | 0.78 | 0.81 |
| INPPL1 | 0.77 | 0.84 |
| ABHD15 | 0.77 | 0.82 |
| CDGAP | 0.77 | 0.76 |
| LRP4 | 0.76 | 0.85 |
| TRIM25 | 0.76 | 0.82 |
| KIAA1161 | 0.76 | 0.76 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NDUFAF2 | -0.50 | -0.56 |
| COMMD3 | -0.48 | -0.52 |
| POLB | -0.47 | -0.51 |
| FRG1 | -0.45 | -0.53 |
| OCIAD2 | -0.45 | -0.48 |
| COQ3 | -0.45 | -0.48 |
| MED19 | -0.44 | -0.47 |
| RPS3AP47 | -0.44 | -0.58 |
| ZNF32 | -0.44 | -0.51 |
| PPP2R3C | -0.44 | -0.49 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME METABOLISM OF PROTEINS | 518 | 242 | All SZGR 2.0 genes in this pathway |
| REACTOME POST TRANSLATIONAL PROTEIN MODIFICATION | 188 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME ASPARAGINE N LINKED GLYCOSYLATION | 81 | 54 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSPORT TO THE GOLGI AND SUBSEQUENT MODIFICATION | 33 | 24 | All SZGR 2.0 genes in this pathway |
| CHEMNITZ RESPONSE TO PROSTAGLANDIN E2 DN | 391 | 222 | All SZGR 2.0 genes in this pathway |
| ZHOU INFLAMMATORY RESPONSE LIVE DN | 384 | 220 | All SZGR 2.0 genes in this pathway |
| CHARAFE BREAST CANCER LUMINAL VS MESENCHYMAL DN | 460 | 312 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| SCHLOSSER SERUM RESPONSE DN | 712 | 443 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS DN | 848 | 527 | All SZGR 2.0 genes in this pathway |
| PASQUALUCCI LYMPHOMA BY GC STAGE UP | 283 | 177 | All SZGR 2.0 genes in this pathway |
| SMITH TERT TARGETS UP | 145 | 91 | All SZGR 2.0 genes in this pathway |
| MANALO HYPOXIA DN | 289 | 166 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| SESTO RESPONSE TO UV C0 | 107 | 72 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL CULTURED VS FRESH UP | 425 | 298 | All SZGR 2.0 genes in this pathway |
| CHNG MULTIPLE MYELOMA HYPERPLOID DN | 28 | 19 | All SZGR 2.0 genes in this pathway |
| ELLWOOD MYC TARGETS DN | 40 | 27 | All SZGR 2.0 genes in this pathway |
| ZHAN LATE DIFFERENTIATION GENES UP | 33 | 24 | All SZGR 2.0 genes in this pathway |
| KYNG WERNER SYNDROM AND NORMAL AGING DN | 225 | 124 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP B | 549 | 316 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS UP | 682 | 433 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |