Gene Page: PAPSS1
Summary ?
| GeneID | 9061 |
| Symbol | PAPSS1 |
| Synonyms | ATPSK1|PAPSS|SK1 |
| Description | 3'-phosphoadenosine 5'-phosphosulfate synthase 1 |
| Reference | MIM:603262|HGNC:HGNC:8603|Ensembl:ENSG00000138801|HPRD:09132|Vega:OTTHUMG00000131210 |
| Gene type | protein-coding |
| Map location | 4q24 |
| Pascal p-value | 0.125 |
| Sherlock p-value | 0.529 |
| Fetal beta | -0.335 |
| DMG | 1 (# studies) |
| eGene | Cerebellum Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg08541219 | 4 | 108641287 | PAPSS1 | -0.03 | 0.32 | DMG:Nishioka_2013 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs2850413 | chr4 | 107959343 | PAPSS1 | 9061 | 0.13 | cis | ||
| rs434649 | chr4 | 107971975 | PAPSS1 | 9061 | 0.12 | cis |
Section II. Transcriptome annotation
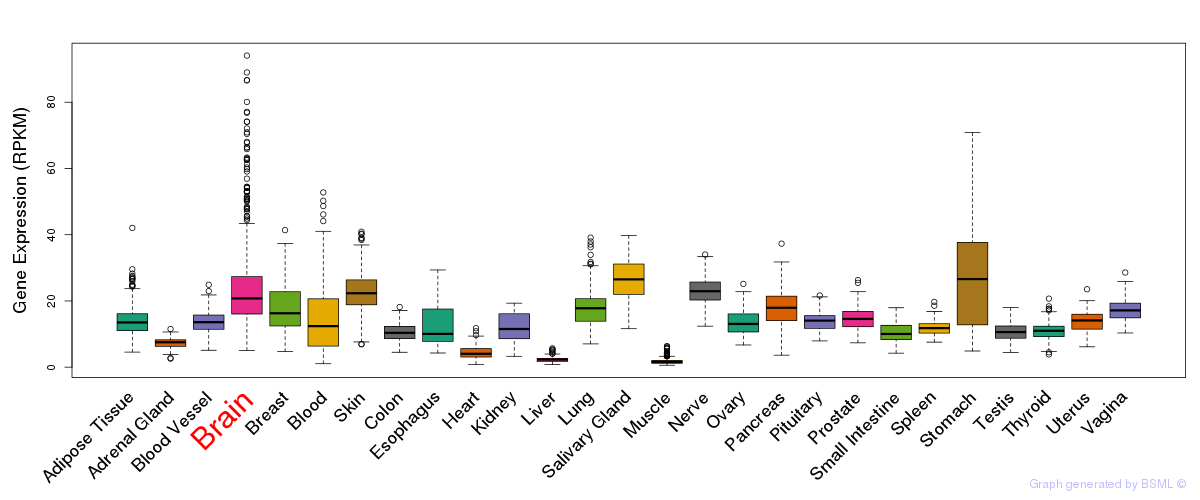
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYNJ1 | 0.94 | 0.90 |
| FAM73A | 0.94 | 0.89 |
| NCOA7 | 0.93 | 0.85 |
| HLF | 0.93 | 0.85 |
| EFR3A | 0.93 | 0.89 |
| GLS | 0.93 | 0.85 |
| MBNL1 | 0.93 | 0.86 |
| UHMK1 | 0.93 | 0.88 |
| ARFGEF2 | 0.93 | 0.82 |
| CHIC1 | 0.93 | 0.81 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.50 | -0.58 |
| FADS2 | -0.46 | -0.32 |
| AC006276.2 | -0.46 | -0.41 |
| NME4 | -0.45 | -0.56 |
| C9orf46 | -0.45 | -0.46 |
| RPL23A | -0.44 | -0.47 |
| TBC1D10A | -0.44 | -0.31 |
| TRAF4 | -0.43 | -0.45 |
| DYNLT1 | -0.43 | -0.52 |
| RPL36 | -0.43 | -0.48 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG PURINE METABOLISM | 159 | 96 | All SZGR 2.0 genes in this pathway |
| KEGG SELENOAMINO ACID METABOLISM | 26 | 18 | All SZGR 2.0 genes in this pathway |
| KEGG SULFUR METABOLISM | 13 | 8 | All SZGR 2.0 genes in this pathway |
| REACTOME BIOLOGICAL OXIDATIONS | 139 | 91 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | 14 | 10 | All SZGR 2.0 genes in this pathway |
| REACTOME PHASE II CONJUGATION | 70 | 42 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DOANE BREAST CANCER CLASSES DN | 34 | 26 | All SZGR 2.0 genes in this pathway |
| HAHTOLA SEZARY SYNDROM UP | 98 | 58 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| LANDIS ERBB2 BREAST TUMORS 324 UP | 150 | 93 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 48HR DN | 428 | 306 | All SZGR 2.0 genes in this pathway |
| PENG GLUCOSE DEPRIVATION DN | 169 | 112 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| REN ALVEOLAR RHABDOMYOSARCOMA DN | 408 | 274 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BAELDE DIABETIC NEPHROPATHY DN | 434 | 302 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 1 | 528 | 324 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER TUMOR VS NORMAL ADJACENT TISSUE UP | 863 | 514 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S1 | 237 | 159 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA UP | 207 | 143 | All SZGR 2.0 genes in this pathway |
| WANG RESPONSE TO GSK3 INHIBITOR SB216763 UP | 397 | 206 | All SZGR 2.0 genes in this pathway |