Gene Page: MAP3K6
Summary ?
| GeneID | 9064 |
| Symbol | MAP3K6 |
| Synonyms | ASK2|MAPKKK6|MEKK6 |
| Description | mitogen-activated protein kinase kinase kinase 6 |
| Reference | MIM:604468|HGNC:HGNC:6858|Ensembl:ENSG00000142733|HPRD:05126|Vega:OTTHUMG00000004631 |
| Gene type | protein-coding |
| Map location | 1p36.11 |
| Pascal p-value | 0.435 |
| Sherlock p-value | 0.839 |
| Fetal beta | -0.629 |
| DMG | 1 (# studies) |
| eGene | Cortex Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg11705208 | 1 | 27683386 | MAP3K6 | 2.39E-5 | 0.379 | 0.017 | DMG:Wockner_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs6753548 | chr2 | 2268763 | MAP3K6 | 9064 | 0.14 | trans | ||
| rs16829545 | chr2 | 151977407 | MAP3K6 | 9064 | 3.549E-5 | trans | ||
| rs16955618 | chr15 | 29937543 | MAP3K6 | 9064 | 2.919E-7 | trans | ||
| rs11873184 | chr18 | 1584081 | MAP3K6 | 9064 | 0.18 | trans |
Section II. Transcriptome annotation
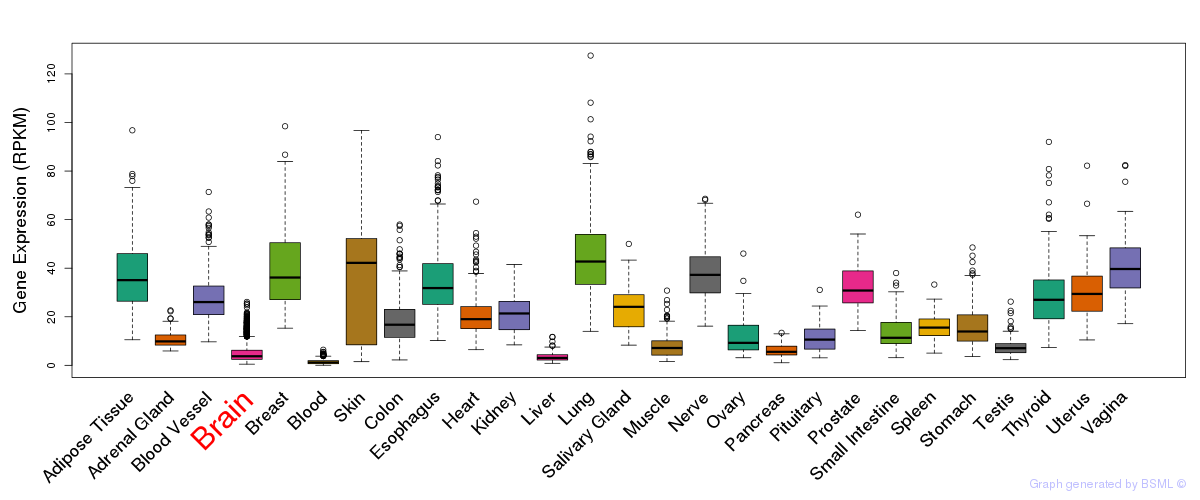
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| NARG1 | 0.96 | 0.97 |
| SERBP1 | 0.95 | 0.94 |
| TOP1 | 0.95 | 0.97 |
| WDR3 | 0.95 | 0.96 |
| SMEK2 | 0.95 | 0.96 |
| SUPT16H | 0.95 | 0.97 |
| ZMYM4 | 0.95 | 0.96 |
| BCLAF1 | 0.95 | 0.97 |
| PPP2R5E | 0.95 | 0.95 |
| CSGALNACT2 | 0.95 | 0.95 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.70 | -0.87 |
| FXYD1 | -0.70 | -0.88 |
| MT-CO2 | -0.69 | -0.88 |
| IFI27 | -0.69 | -0.87 |
| HLA-F | -0.69 | -0.76 |
| TSC22D4 | -0.68 | -0.80 |
| C5orf53 | -0.68 | -0.73 |
| AF347015.27 | -0.68 | -0.84 |
| AIFM3 | -0.68 | -0.76 |
| AF347015.33 | -0.68 | -0.83 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MAPK SIGNALING PATHWAY | 267 | 205 | All SZGR 2.0 genes in this pathway |
| BIOCARTA MAPK PATHWAY | 87 | 68 | All SZGR 2.0 genes in this pathway |
| PID P38 MKK3 6PATHWAY | 26 | 21 | All SZGR 2.0 genes in this pathway |
| PARENT MTOR SIGNALING UP | 567 | 375 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER UP | 443 | 294 | All SZGR 2.0 genes in this pathway |
| SCHAEFFER PROSTATE DEVELOPMENT 12HR UP | 116 | 79 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 2 | 72 | 52 | All SZGR 2.0 genes in this pathway |
| BILD MYC ONCOGENIC SIGNATURE | 206 | 117 | All SZGR 2.0 genes in this pathway |
| HELLER SILENCED BY METHYLATION UP | 282 | 183 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO CSF2RB AND IL4 UP | 338 | 225 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF DN | 235 | 144 | All SZGR 2.0 genes in this pathway |
| RUTELLA RESPONSE TO HGF VS CSF2RB AND IL4 DN | 245 | 150 | All SZGR 2.0 genes in this pathway |
| VANASSE BCL2 TARGETS UP | 40 | 25 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER GOOD SURVIVAL A4 | 196 | 124 | All SZGR 2.0 genes in this pathway |
| MEISSNER BRAIN HCP WITH H3K4ME3 AND H3K27ME3 | 1069 | 729 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |
| DURAND STROMA NS UP | 162 | 103 | All SZGR 2.0 genes in this pathway |