Gene Page: USP8
Summary ?
| GeneID | 9101 |
| Symbol | USP8 |
| Synonyms | HumORF8|SPG59|UBPY |
| Description | ubiquitin specific peptidase 8 |
| Reference | MIM:603158|HGNC:HGNC:12631|HPRD:04405| |
| Gene type | protein-coding |
| Map location | 15q21.2 |
| Pascal p-value | 0.884 |
| Sherlock p-value | 0.12 |
| Fetal beta | -0.968 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Nishioka_2013 | Genome-wide DNA methylation analysis | The authors investigated the methylation profiles of DNA in peripheral blood cells from 18 patients with first-episode schizophrenia (FESZ) and from 15 normal controls. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg01443067 | 15 | 50716755 | USP8 | -0.024 | 0.44 | DMG:Nishioka_2013 |
Section II. Transcriptome annotation
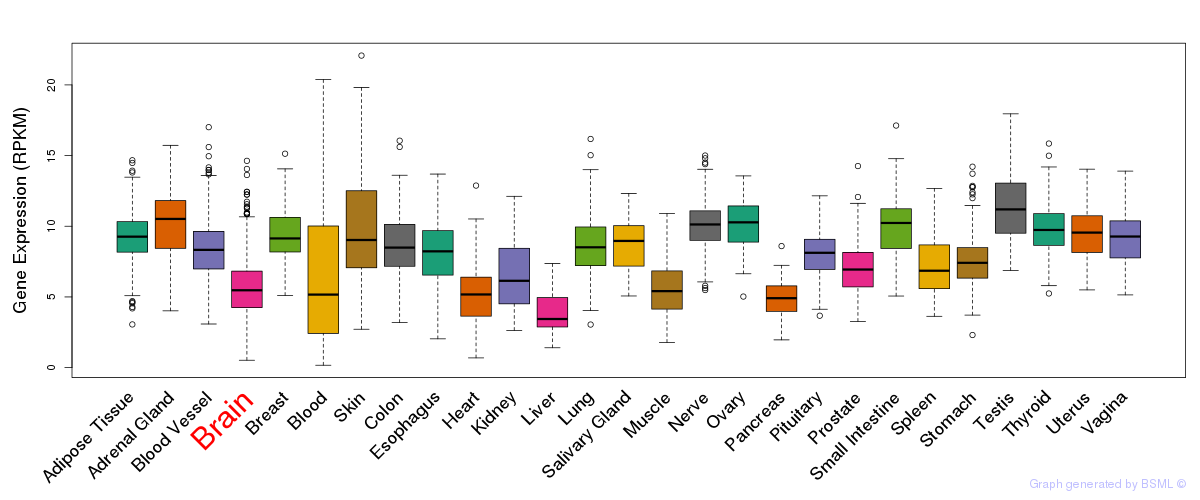
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| GRB2 | ASH | EGFRBP-GRB2 | Grb3-3 | MST084 | MSTP084 | growth factor receptor-bound protein 2 | - | HPRD,BioGRID | 10982817 |13129930 |
| OTUB1 | FLJ20113 | FLJ40710 | HSPC263 | MGC111158 | MGC4584 | OTB1 | OTU1 | OTU domain, ubiquitin aldehyde binding 1 | Affinity Capture-Western | BioGRID | 14661020 |
| RASGRF1 | CDC25 | CDC25L | GNRP | GRF1 | GRF55 | H-GRF55 | PP13187 | Ras protein-specific guanine nucleotide-releasing factor 1 | - | HPRD | 11500497 |
| RNF128 | FLJ23516 | GRAIL | ring finger protein 128 | Affinity Capture-Western | BioGRID | 14661020 |
| RNF41 | MGC45228 | NRDP1 | SBBI03 | ring finger protein 41 | - | HPRD,BioGRID | 15314180 |
| SFN | YWHAS | stratifin | Affinity Capture-MS | BioGRID | 15778465 |
| SLBP | HBP | stem-loop binding protein | - | HPRD | 10982817 |
| STAM | DKFZp686J2352 | STAM1 | signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | - | HPRD,BioGRID | 10982817 |
| STAM2 | DKFZp564C047 | Hbp | STAM2A | STAM2B | signal transducing adaptor molecule (SH3 domain and ITAM motif) 2 | - | HPRD,BioGRID | 13129930 |
| YWHAB | GW128 | HS1 | KCIP-1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAQ | 14-3-3 | 1C5 | HS1 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, theta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
| YWHAZ | KCIP-1 | MGC111427 | MGC126532 | MGC138156 | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG ENDOCYTOSIS | 183 | 132 | All SZGR 2.0 genes in this pathway |
| PID ERBB2 ERBB3 PATHWAY | 44 | 35 | All SZGR 2.0 genes in this pathway |
| PID ERBB1 INTERNALIZATION PATHWAY | 41 | 35 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | 12 | 9 | All SZGR 2.0 genes in this pathway |
| REACTOME SIGNALING BY ERBB2 | 101 | 78 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS DN | 352 | 225 | All SZGR 2.0 genes in this pathway |
| HAMAI APOPTOSIS VIA TRAIL UP | 584 | 356 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| GOTTWEIN TARGETS OF KSHV MIR K12 11 | 63 | 45 | All SZGR 2.0 genes in this pathway |
| YAGI AML FAB MARKERS | 191 | 131 | All SZGR 2.0 genes in this pathway |
| URS ADIPOCYTE DIFFERENTIATION UP | 74 | 51 | All SZGR 2.0 genes in this pathway |
| YIH RESPONSE TO ARSENITE C2 | 18 | 15 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| WONG PROTEASOME GENE MODULE | 49 | 35 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER UP | 973 | 570 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH INV 16 TRANSLOCATION | 422 | 277 | All SZGR 2.0 genes in this pathway |
| ASGHARZADEH NEUROBLASTOMA POOR SURVIVAL DN | 46 | 30 | All SZGR 2.0 genes in this pathway |
| SAKAI CHRONIC HEPATITIS VS LIVER CANCER UP | 83 | 63 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS UP | 504 | 321 | All SZGR 2.0 genes in this pathway |
| JUBAN TARGETS OF SPI1 AND FLI1 UP | 115 | 73 | All SZGR 2.0 genes in this pathway |