Gene Page: ATG12
Summary ?
| GeneID | 9140 |
| Symbol | ATG12 |
| Synonyms | APG12|APG12L|FBR93|HAPG12 |
| Description | autophagy related 12 |
| Reference | MIM:609608|HGNC:HGNC:588|Ensembl:ENSG00000145782|HPRD:08496|Vega:OTTHUMG00000128889 |
| Gene type | protein-coding |
| Map location | 5q21-q22 |
| Pascal p-value | 0.108 |
| TADA p-value | 5.53E-4 |
| Fetal beta | 1.262 |
| eGene | Caudate basal ganglia Cerebellum Myers' cis & trans |
| Support | CompositeSet |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ATG12 | chr5 | 115173428..115173432 | GGAGT | G | NM_001277783 NM_004707 NR_033362 NR_033363 NR_073603 NR_073604 NR_073605 | . . . . . . . | intronic frameshift npcRNA npcRNA npcRNA intronic intronic | Schizophrenia | DNM:Fromer_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs4366501 | chr11 | 133380324 | ATG12 | 9140 | 0.01 | trans |
Section II. Transcriptome annotation
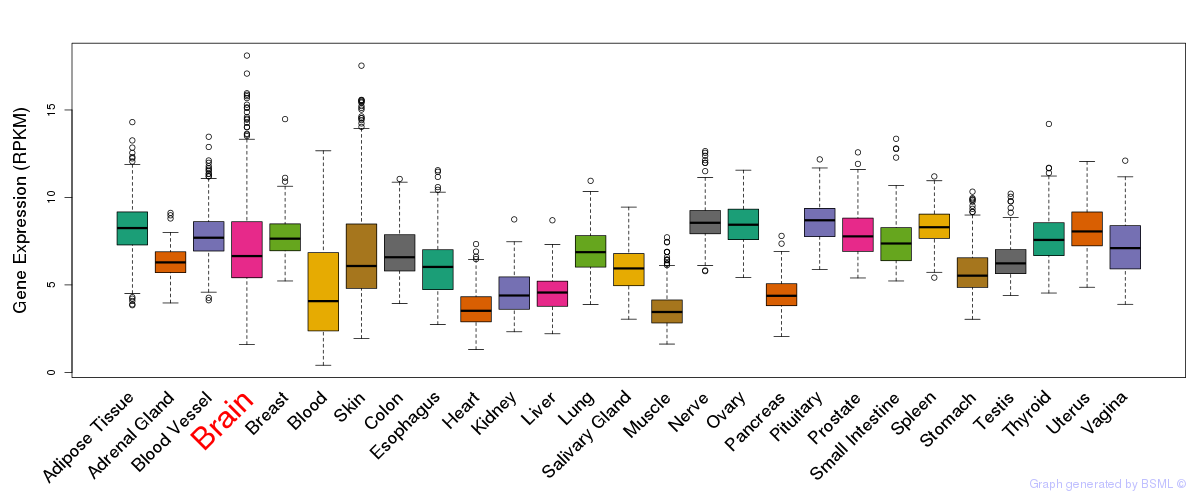
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| UROS | 0.82 | 0.72 |
| NDUFB6 | 0.79 | 0.72 |
| C1orf31 | 0.78 | 0.77 |
| HINT1 | 0.77 | 0.71 |
| TXNL4A | 0.76 | 0.70 |
| CALM2 | 0.75 | 0.83 |
| PTS | 0.75 | 0.78 |
| THOC7 | 0.75 | 0.75 |
| C6orf57 | 0.75 | 0.68 |
| UQCRFSL1 | 0.75 | 0.69 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AC020763.2 | -0.43 | -0.45 |
| CASKIN2 | -0.41 | -0.45 |
| MTSS1L | -0.41 | -0.40 |
| FAM59B | -0.40 | -0.44 |
| EPHA2 | -0.40 | -0.43 |
| CDC42EP4 | -0.40 | -0.44 |
| SMTN | -0.39 | -0.46 |
| AF347015.18 | -0.38 | -0.21 |
| PLXNB1 | -0.38 | -0.43 |
| TENC1 | -0.38 | -0.40 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATG10 | APG10 | APG10L | DKFZp586I0418 | FLJ13954 | pp12616 | ATG10 autophagy related 10 homolog (S. cerevisiae) | - | HPRD | 12482611 |
| ATG10 | APG10 | APG10L | DKFZp586I0418 | FLJ13954 | pp12616 | ATG10 autophagy related 10 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| ATG16L1 | APG16L | ATG16L | FLJ00045 | FLJ10035 | FLJ10828 | FLJ22677 | IBD10 | WDR30 | ATG16 autophagy related 16-like 1 (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| ATG3 | APG3 | APG3-LIKE | APG3L | DKFZp564M1178 | FLJ22125 | MGC15201 | PC3-96 | ATG3 autophagy related 3 homolog (S. cerevisiae) | - | HPRD,BioGRID | 11825910 |
| ATG5 | APG5 | APG5-LIKE | APG5L | ASP | hAPG5 | ATG5 autophagy related 5 homolog (S. cerevisiae) | - | HPRD | 11825910 |12482611 |
| ATG5 | APG5 | APG5-LIKE | APG5L | ASP | hAPG5 | ATG5 autophagy related 5 homolog (S. cerevisiae) | Affinity Capture-MS | BioGRID | 17353931 |
| ATG7 | APG7-LIKE | APG7L | DKFZp434N0735 | GSA7 | ATG7 autophagy related 7 homolog (S. cerevisiae) | - | HPRD | 11890701 |12482611 |
| AUP1 | - | ancient ubiquitous protein 1 | Affinity Capture-MS | BioGRID | 17353931 |
| DHX36 | DDX36 | G4R1 | KIAA1488 | MLEL1 | RHAU | DEAH (Asp-Glu-Ala-His) box polypeptide 36 | Affinity Capture-MS | BioGRID | 17353931 |
| KRTAP4-12 | KAP4.12 | KRTAP4.12 | keratin associated protein 4-12 | Two-hybrid | BioGRID | 16189514 |
| MDFI | I-MF | I-mfa | MyoD family inhibitor | Two-hybrid | BioGRID | 16189514 |
| OTUD4 | DKFZp434I0721 | DUBA6 | HIN1 | HSHIN1 | KIAA1046 | OTU domain containing 4 | Affinity Capture-MS | BioGRID | 17353931 |
| PLSCR1 | MMTRA1B | phospholipid scramblase 1 | Two-hybrid | BioGRID | 16189514 |
| PTK2 | FADK | FAK | FAK1 | pp125FAK | PTK2 protein tyrosine kinase 2 | Two-hybrid | BioGRID | 16189514 |
| SF3A1 | PRP21 | PRPF21 | SAP114 | SF3A120 | splicing factor 3a, subunit 1, 120kDa | Affinity Capture-MS | BioGRID | 17353931 |
| SF3B1 | PRP10 | PRPF10 | SAP155 | SF3b155 | splicing factor 3b, subunit 1, 155kDa | Affinity Capture-MS | BioGRID | 17353931 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG REGULATION OF AUTOPHAGY | 35 | 30 | All SZGR 2.0 genes in this pathway |
| KEGG RIG I LIKE RECEPTOR SIGNALING PATHWAY | 71 | 51 | All SZGR 2.0 genes in this pathway |
| REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | 31 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME RIG I MDA5 MEDIATED INDUCTION OF IFN ALPHA BETA PATHWAYS | 73 | 57 | All SZGR 2.0 genes in this pathway |
| REACTOME INNATE IMMUNE SYSTEM | 279 | 178 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN | 92 | 51 | All SZGR 2.0 genes in this pathway |
| PUIFFE INVASION INHIBITED BY ASCITES DN | 145 | 91 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| WANG CLIM2 TARGETS DN | 186 | 114 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED UP | 78 | 48 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA ANAPLASTIC UP | 722 | 443 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| HAHTOLA MYCOSIS FUNGOIDES CD4 DN | 116 | 71 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| BARRIER CANCER RELAPSE TUMOR SAMPLE UP | 16 | 12 | All SZGR 2.0 genes in this pathway |
| BERENJENO TRANSFORMED BY RHOA DN | 394 | 258 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| FLECHNER BIOPSY KIDNEY TRANSPLANT OK VS DONOR UP | 555 | 346 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 STIMULATED | 1022 | 619 | All SZGR 2.0 genes in this pathway |
| MARSON BOUND BY FOXP3 UNSTIMULATED | 1229 | 713 | All SZGR 2.0 genes in this pathway |
| HELLER HDAC TARGETS SILENCED BY METHYLATION UP | 461 | 298 | All SZGR 2.0 genes in this pathway |
| MITSIADES RESPONSE TO APLIDIN UP | 439 | 257 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS | 425 | 261 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
| WAKABAYASHI ADIPOGENESIS PPARG RXRA BOUND 8D | 882 | 506 | All SZGR 2.0 genes in this pathway |
| MIZUSHIMA AUTOPHAGOSOME FORMATION | 19 | 15 | All SZGR 2.0 genes in this pathway |