Gene Page: CD247
Summary ?
| GeneID | 919 |
| Symbol | CD247 |
| Synonyms | CD3-ZETA|CD3H|CD3Q|CD3Z|IMD25|T3Z|TCRZ |
| Description | CD247 molecule |
| Reference | MIM:186780|HGNC:HGNC:1677|Ensembl:ENSG00000198821|HPRD:01729|Vega:OTTHUMG00000034593 |
| Gene type | protein-coding |
| Map location | 1q24.2 |
| Sherlock p-value | 0.696 |
| Fetal beta | -0.317 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg09554443 | 1 | 167487762 | CD247 | 0.004 | -4.422 | DMG:vanEijk_2014 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs13375406 | chr1 | 95606304 | CD247 | 919 | 0.13 | trans | ||
| rs16838771 | chr1 | 157511227 | CD247 | 919 | 7.906E-5 | trans | ||
| rs16856639 | chr1 | 166116296 | CD247 | 919 | 0.15 | trans | ||
| rs973381 | chr1 | 166121317 | CD247 | 919 | 0.15 | trans | ||
| rs1538755 | chr1 | 210893874 | CD247 | 919 | 0.13 | trans | ||
| rs825935 | chr2 | 4808721 | CD247 | 919 | 0.19 | trans | ||
| rs825932 | chr2 | 4811625 | CD247 | 919 | 0.07 | trans | ||
| rs16865228 | chr2 | 6629622 | CD247 | 919 | 0.1 | trans | ||
| rs16865230 | chr2 | 6630187 | CD247 | 919 | 0.1 | trans | ||
| rs11691188 | chr2 | 6631799 | CD247 | 919 | 0.1 | trans | ||
| rs3755119 | chr2 | 53957339 | CD247 | 919 | 0.05 | trans | ||
| rs17045286 | chr2 | 54049393 | CD247 | 919 | 3.687E-4 | trans | ||
| rs10202944 | chr2 | 72257857 | CD247 | 919 | 0.15 | trans | ||
| rs7602648 | chr2 | 104661176 | CD247 | 919 | 0.03 | trans | ||
| rs6708612 | chr2 | 148165722 | CD247 | 919 | 0.17 | trans | ||
| rs6729020 | chr2 | 189064719 | CD247 | 919 | 0.13 | trans | ||
| snp_a-2186147 | 0 | CD247 | 919 | 0.02 | trans | |||
| rs6803351 | chr3 | 17336986 | CD247 | 919 | 0 | trans | ||
| rs11916205 | chr3 | 17392363 | CD247 | 919 | 0 | trans | ||
| rs17043486 | chr3 | 17399005 | CD247 | 919 | 0 | trans | ||
| rs17029249 | chr3 | 17416384 | CD247 | 919 | 0 | trans | ||
| rs10510474 | chr3 | 17417495 | CD247 | 919 | 0 | trans | ||
| rs17043623 | chr3 | 17478890 | CD247 | 919 | 0 | trans | ||
| rs11128830 | chr3 | 17481727 | CD247 | 919 | 0 | trans | ||
| rs4378948 | 0 | CD247 | 919 | 0.12 | trans | |||
| rs11927611 | chr3 | 17848434 | CD247 | 919 | 0 | trans | ||
| rs6793958 | chr3 | 17853055 | CD247 | 919 | 2.217E-10 | trans | ||
| rs17044072 | chr3 | 17854496 | CD247 | 919 | 0.01 | trans | ||
| rs17044097 | chr3 | 17889750 | CD247 | 919 | 0 | trans | ||
| rs17044102 | chr3 | 17889838 | CD247 | 919 | 0.03 | trans | ||
| rs4591529 | chr3 | 32330610 | CD247 | 919 | 0.15 | trans | ||
| rs7626782 | 0 | CD247 | 919 | 0.03 | trans | |||
| rs2600956 | chr3 | 132624112 | CD247 | 919 | 0.12 | trans | ||
| rs1452144 | chr3 | 132627627 | CD247 | 919 | 0.12 | trans | ||
| rs7635430 | chr3 | 180152035 | CD247 | 919 | 0.11 | trans | ||
| rs1291259 | 0 | CD247 | 919 | 0 | trans | |||
| rs17005625 | chr4 | 141141923 | CD247 | 919 | 0.09 | trans | ||
| rs17005641 | chr4 | 141152656 | CD247 | 919 | 0.04 | trans | ||
| rs10026641 | chr4 | 183431866 | CD247 | 919 | 0 | trans | ||
| rs11723043 | chr4 | 189507117 | CD247 | 919 | 0.04 | trans | ||
| rs4702089 | chr5 | 15640330 | CD247 | 919 | 0.08 | trans | ||
| rs4702104 | chr5 | 15746397 | CD247 | 919 | 0.01 | trans | ||
| rs17138463 | chr5 | 115286058 | CD247 | 919 | 0.07 | trans | ||
| rs17138468 | chr5 | 115288423 | CD247 | 919 | 0.05 | trans | ||
| rs10519531 | chr5 | 117332352 | CD247 | 919 | 2.702E-10 | trans | ||
| rs6867333 | chr5 | 117339065 | CD247 | 919 | 0.05 | trans | ||
| rs17736846 | chr5 | 169133824 | CD247 | 919 | 0.02 | trans | ||
| rs9503329 | chr6 | 2894607 | CD247 | 919 | 0 | trans | ||
| rs17458000 | chr6 | 151384414 | CD247 | 919 | 0 | trans | ||
| rs1321606 | chr6 | 153545278 | CD247 | 919 | 0.17 | trans | ||
| rs163007 | chr6 | 155726495 | CD247 | 919 | 0.15 | trans | ||
| rs7760005 | chr6 | 156850663 | CD247 | 919 | 0.02 | trans | ||
| rs11983989 | chr7 | 46351006 | CD247 | 919 | 0.09 | trans | ||
| rs17172680 | chr7 | 46362331 | CD247 | 919 | 0.09 | trans | ||
| rs17172688 | chr7 | 46396563 | CD247 | 919 | 0.09 | trans | ||
| rs11978272 | chr7 | 80119689 | CD247 | 919 | 0.2 | trans | ||
| rs17163613 | chr7 | 90764256 | CD247 | 919 | 6.475E-5 | trans | ||
| rs17684427 | chr7 | 122482976 | CD247 | 919 | 0.15 | trans | ||
| rs7796458 | chr7 | 154873701 | CD247 | 919 | 0.13 | trans | ||
| rs1474448 | chr8 | 16353122 | CD247 | 919 | 0.09 | trans | ||
| rs4737205 | chr8 | 65995897 | CD247 | 919 | 0.19 | trans | ||
| rs17495371 | 0 | CD247 | 919 | 0 | trans | |||
| rs17495754 | chr8 | 82152549 | CD247 | 919 | 0.08 | trans | ||
| rs7813477 | chr8 | 85144311 | CD247 | 919 | 0.06 | trans | ||
| rs4878586 | chr9 | 27810413 | CD247 | 919 | 0.05 | trans | ||
| rs1410577 | chr9 | 90283060 | CD247 | 919 | 0.18 | trans | ||
| rs4742740 | chr9 | 101524552 | CD247 | 919 | 0.07 | trans | ||
| rs4743277 | chr9 | 101557528 | CD247 | 919 | 0.06 | trans | ||
| rs11252838 | chr10 | 4972173 | CD247 | 919 | 0.01 | trans | ||
| rs16933426 | chr10 | 78113005 | CD247 | 919 | 2.036E-5 | trans | ||
| rs16933436 | chr10 | 78118247 | CD247 | 919 | 0.18 | trans | ||
| rs16933437 | chr10 | 78118689 | CD247 | 919 | 0.18 | trans | ||
| rs16933467 | chr10 | 78141332 | CD247 | 919 | 0.18 | trans | ||
| rs10885916 | chr10 | 118118819 | CD247 | 919 | 0.04 | trans | ||
| rs11199540 | chr10 | 122530843 | CD247 | 919 | 0.02 | trans | ||
| rs2459087 | chr10 | 123821264 | CD247 | 919 | 0.01 | trans | ||
| rs11016704 | chr10 | 128989145 | CD247 | 919 | 0.17 | trans | ||
| rs6486418 | 0 | CD247 | 919 | 3.568E-4 | trans | |||
| rs12292520 | chr11 | 48086150 | CD247 | 919 | 0.05 | trans | ||
| rs17789481 | chr11 | 48110042 | CD247 | 919 | 0.2 | trans | ||
| rs11220020 | chr11 | 125183479 | CD247 | 919 | 0.09 | trans | ||
| rs10878919 | chr12 | 69521979 | CD247 | 919 | 5.246E-4 | trans | ||
| rs7136572 | chr12 | 95959787 | CD247 | 919 | 6.866E-9 | trans | ||
| rs935031 | chr12 | 96861369 | CD247 | 919 | 0.12 | trans | ||
| rs10848152 | chr12 | 131062467 | CD247 | 919 | 0 | trans | ||
| rs11061005 | chr12 | 131063527 | CD247 | 919 | 0 | trans | ||
| rs7989474 | chr13 | 29113187 | CD247 | 919 | 0.16 | trans | ||
| rs1335797 | chr13 | 64425435 | CD247 | 919 | 1.019E-5 | trans | ||
| rs17115591 | chr14 | 45262026 | CD247 | 919 | 0.13 | trans | ||
| rs12588364 | chr14 | 79411335 | CD247 | 919 | 0.12 | trans | ||
| rs11159388 | chr14 | 79417713 | CD247 | 919 | 0.12 | trans | ||
| rs11159390 | chr14 | 79424474 | CD247 | 919 | 0.12 | trans | ||
| rs10147554 | chr14 | 89902966 | CD247 | 919 | 0.13 | trans | ||
| rs11623367 | chr14 | 89924832 | CD247 | 919 | 0 | trans | ||
| rs8026736 | chr15 | 45364871 | CD247 | 919 | 0.14 | trans | ||
| rs10518880 | chr15 | 70491974 | CD247 | 919 | 0.02 | trans | ||
| rs7180781 | chr15 | 78455354 | CD247 | 919 | 0.13 | trans | ||
| rs17135416 | chr16 | 1645744 | CD247 | 919 | 0.17 | trans | ||
| rs4780835 | chr16 | 19985287 | CD247 | 919 | 0.12 | trans | ||
| rs7198149 | chr16 | 57920460 | CD247 | 919 | 0.03 | trans | ||
| rs7197514 | chr16 | 65652629 | CD247 | 919 | 0.01 | trans | ||
| rs16944062 | chr16 | 76272616 | CD247 | 919 | 0.01 | trans | ||
| rs16942128 | chr17 | 45764234 | CD247 | 919 | 0.15 | trans | ||
| rs12968895 | chr18 | 19650706 | CD247 | 919 | 0.13 | trans | ||
| rs9304469 | chr18 | 22169423 | CD247 | 919 | 0.06 | trans | ||
| rs17187821 | chr18 | 22209158 | CD247 | 919 | 0.05 | trans | ||
| rs17785402 | chr20 | 5173471 | CD247 | 919 | 0.06 | trans | ||
| rs2830443 | chr21 | 28109967 | CD247 | 919 | 0.16 | trans | ||
| rs6586271 | chr21 | 44362641 | CD247 | 919 | 0.03 | trans | ||
| rs12627758 | chr22 | 29382830 | CD247 | 919 | 0.03 | trans | ||
| rs16989337 | chr22 | 32025054 | CD247 | 919 | 0.13 | trans |
Section II. Transcriptome annotation
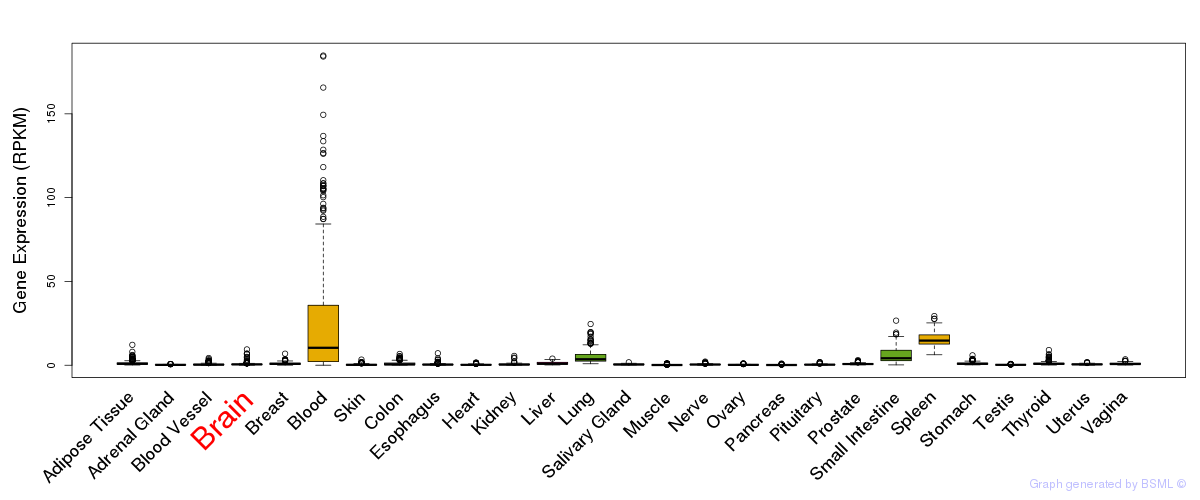
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SECTM1 | 0.64 | 0.27 |
| C19orf35 | 0.63 | 0.43 |
| KISS1R | 0.62 | 0.21 |
| PTGER1 | 0.57 | 0.42 |
| SMOC2 | 0.56 | 0.54 |
| AC011500.2 | 0.54 | 0.31 |
| C1orf133 | 0.54 | 0.39 |
| P2RY1 | 0.54 | 0.27 |
| RXRG | 0.52 | 0.33 |
| FAM40B | 0.51 | 0.36 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| C18orf8 | -0.44 | -0.58 |
| GTDC1 | -0.43 | -0.47 |
| GARNL3 | -0.42 | -0.52 |
| CABYR | -0.41 | -0.58 |
| ZNF30 | -0.40 | -0.56 |
| CBARA1 | -0.40 | -0.53 |
| SFXN3 | -0.40 | -0.51 |
| DYNC1I1 | -0.39 | -0.48 |
| RNF8 | -0.39 | -0.51 |
| RAD51C | -0.39 | -0.42 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0004888 | transmembrane receptor activity | IEA | - | |
| GO:0005515 | protein binding | IEA | - | |
| GO:0042803 | protein homodimerization activity | NAS | 12110186 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007166 | cell surface receptor linked signal transduction | IEA | - | |
| GO:0044419 | interspecies interaction between organisms | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0042101 | T cell receptor complex | IDA | Synap (GO term level: 8) | 8176201 |
| GO:0005737 | cytoplasm | IDA | 11390434 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0016021 | integral to membrane | IEA | - | |
| GO:0005886 | plasma membrane | EXP | 11048639 |11827988 |15489916 |17652306 | |
| GO:0005886 | plasma membrane | IDA | 1390434 |11390434 | |
| GO:0042105 | alpha-beta T cell receptor complex | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ATXN2 | ATX2 | FLJ46772 | SCA2 | TNRC13 | ataxin 2 | Affinity Capture-Western | BioGRID | 9575182 |
| CD2 | SRBC | T11 | CD2 molecule | - | HPRD | 8977276 |
| CD247 | CD3-ZETA | CD3H | CD3Q | CD3Z | T3Z | TCRZ | CD247 molecule | - | HPRD | 2845582 |
| CD28 | MGC138290 | Tp44 | CD28 molecule | - | HPRD | 8760790 |
| CD3D | CD3-DELTA | T3D | CD3d molecule, delta (CD3-TCR complex) | - | HPRD | 9485181 |
| CD3E | FLJ18683 | T3E | TCRE | CD3e molecule, epsilon (CD3-TCR complex) | - | HPRD | 9485181 |
| CD5 | LEU1 | T1 | CD5 molecule | - | HPRD | 1385158 |
| CSK | MGC117393 | c-src tyrosine kinase | - | HPRD | 9172452 |
| DOCK2 | FLJ46592 | KIAA0209 | dedicator of cytokinesis 2 | - | HPRD,BioGRID | 12176041 |
| GAB2 | KIAA0571 | GRB2-associated binding protein 2 | - | HPRD,BioGRID | 11572860 |
| JAK3 | JAK-3 | JAK3_HUMAN | JAKL | L-JAK | LJAK | Janus kinase 3 (a protein tyrosine kinase, leukocyte) | - | HPRD,BioGRID | 11349123 |
| LY6E | RIG-E | RIGE | SCA-2 | SCA2 | TSA-1 | lymphocyte antigen 6 complex, locus E | - | HPRD | 9575182 |
| NCR1 | CD335 | LY94 | NK-p46 | NKP46 | natural cytotoxicity triggering receptor 1 | - | HPRD,BioGRID | 9625766 |
| NCR3 | 1C7 | CD337 | LY117 | MALS | NKp30 | natural cytotoxicity triggering receptor 3 | - | HPRD | 12731048 |
| PTPRC | B220 | CD45 | CD45R | GP180 | LCA | LY5 | T200 | protein tyrosine phosphatase, receptor type, C | - | HPRD,BioGRID | 7526385 |
| SH2B3 | CELIAC13 | LNK | SH2B adaptor protein 3 | - | HPRD | 10799879 |
| SHC1 | FLJ26504 | SHC | SHCA | SHC (Src homology 2 domain containing) transforming protein 1 | - | HPRD | 8656061 |
| SLA | SLA1 | SLAP | Src-like-adaptor | - | HPRD,BioGRID | 10449770 |
| SLA2 | C20orf156 | FLJ21992 | MGC49845 | SLAP-2 | SLAP2 | Src-like-adaptor 2 | - | HPRD,BioGRID | 11891219 |
| STAT5A | MGF | STAT5 | signal transducer and activator of transcription 5A | - | HPRD,BioGRID | 9880255 |
| STAT5B | STAT5 | signal transducer and activator of transcription 5B | - | HPRD,BioGRID | 9880255 |
| TFRC | CD71 | TFR | TFR1 | TRFR | transferrin receptor (p90, CD71) | - | HPRD | 9448136 |
| TIRAP | FLJ42305 | Mal | wyatt | toll-interleukin 1 receptor (TIR) domain containing adaptor protein | - | HPRD | 11572860 |
| TRA@ | FLJ22602 | MGC117436 | MGC22624 | MGC23964 | MGC71411 | TCRA | TCRD | TRA | T cell receptor alpha locus | - | HPRD | 2138083 |
| TRAT1 | HSPC062 | TCRIM | TRIM | T cell receptor associated transmembrane adaptor 1 | - | HPRD | 9687533 |11390434 |
| TRAT1 | HSPC062 | TCRIM | TRIM | T cell receptor associated transmembrane adaptor 1 | - | HPRD,BioGRID | 11390434 |
| UNC119 | HRG4 | unc-119 homolog (C. elegans) | - | HPRD,BioGRID | 14757743 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | - | HPRD | 8366117 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG NATURAL KILLER CELL MEDIATED CYTOTOXICITY | 137 | 92 | All SZGR 2.0 genes in this pathway |
| KEGG T CELL RECEPTOR SIGNALING PATHWAY | 108 | 89 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CSK PATHWAY | 24 | 20 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTL PATHWAY | 15 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCAPOPTOSIS PATHWAY | 11 | 8 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL17 PATHWAY | 17 | 12 | All SZGR 2.0 genes in this pathway |
| BIOCARTA IL12 PATHWAY | 23 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCRA PATHWAY | 13 | 10 | All SZGR 2.0 genes in this pathway |
| BIOCARTA NO2IL12 PATHWAY | 17 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TOB1 PATHWAY | 21 | 14 | All SZGR 2.0 genes in this pathway |
| BIOCARTA STATHMIN PATHWAY | 19 | 17 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCR PATHWAY | 49 | 37 | All SZGR 2.0 genes in this pathway |
| BIOCARTA TCYTOTOXIC PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA THELPER PATHWAY | 14 | 11 | All SZGR 2.0 genes in this pathway |
| BIOCARTA CTLA4 PATHWAY | 21 | 18 | All SZGR 2.0 genes in this pathway |
| PID TCR PATHWAY | 66 | 51 | All SZGR 2.0 genes in this pathway |
| PID IL12 2PATHWAY | 63 | 54 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR PATHWAY | 53 | 42 | All SZGR 2.0 genes in this pathway |
| PID CXCR4 PATHWAY | 102 | 78 | All SZGR 2.0 genes in this pathway |
| PID HIV NEF PATHWAY | 35 | 26 | All SZGR 2.0 genes in this pathway |
| PID CD8 TCR DOWNSTREAM PATHWAY | 65 | 56 | All SZGR 2.0 genes in this pathway |
| PID IL12 STAT4 PATHWAY | 33 | 29 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNOREGULATORY INTERACTIONS BETWEEN A LYMPHOID AND A NON LYMPHOID CELL | 70 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME TCR SIGNALING | 54 | 46 | All SZGR 2.0 genes in this pathway |
| REACTOME DOWNSTREAM TCR SIGNALING | 37 | 32 | All SZGR 2.0 genes in this pathway |
| REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | 16 | 15 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | 14 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERATION OF SECOND MESSENGER MOLECULES | 27 | 25 | All SZGR 2.0 genes in this pathway |
| REACTOME PD1 SIGNALING | 18 | 14 | All SZGR 2.0 genes in this pathway |
| REACTOME COSTIMULATION BY THE CD28 FAMILY | 63 | 48 | All SZGR 2.0 genes in this pathway |
| REACTOME HIV INFECTION | 207 | 122 | All SZGR 2.0 genes in this pathway |
| REACTOME HOST INTERACTIONS OF HIV FACTORS | 132 | 81 | All SZGR 2.0 genes in this pathway |
| REACTOME THE ROLE OF NEF IN HIV1 REPLICATION AND DISEASE PATHOGENESIS | 28 | 19 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME ADAPTIVE IMMUNE SYSTEM | 539 | 350 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| MATSUDA NATURAL KILLER DIFFERENTIATION | 475 | 313 | All SZGR 2.0 genes in this pathway |
| MOREIRA RESPONSE TO TSA DN | 18 | 16 | All SZGR 2.0 genes in this pathway |
| SCHLINGEMANN SKIN CARCINOGENESIS TPA DN | 29 | 15 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP | 202 | 115 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER LUMINAL B DN | 564 | 326 | All SZGR 2.0 genes in this pathway |
| SMID BREAST CANCER NORMAL LIKE UP | 476 | 285 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| LEE DIFFERENTIATING T LYMPHOCYTE | 200 | 115 | All SZGR 2.0 genes in this pathway |
| FINAK BREAST CANCER SDPP SIGNATURE | 26 | 11 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| SENGUPTA EBNA1 ANTICORRELATED | 173 | 85 | All SZGR 2.0 genes in this pathway |
| BRUINS UVC RESPONSE VIA TP53 GROUP A | 898 | 516 | All SZGR 2.0 genes in this pathway |
| KASLER HDAC7 TARGETS 1 UP | 194 | 133 | All SZGR 2.0 genes in this pathway |
| ROESSLER LIVER CANCER METASTASIS UP | 107 | 72 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-214 | 410 | 416 | m8 | hsa-miR-214brain | ACAGCAGGCACAGACAGGCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.