Gene Page: CD5
Summary ?
| GeneID | 921 |
| Symbol | CD5 |
| Synonyms | LEU1|T1 |
| Description | CD5 molecule |
| Reference | MIM:153340|HGNC:HGNC:1685|Ensembl:ENSG00000110448|HPRD:01078|Vega:OTTHUMG00000167825 |
| Gene type | protein-coding |
| Map location | 11q13 |
| Pascal p-value | 0.464 |
| Fetal beta | -0.062 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum Cortex |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 1 |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenic,schizophrenics,schizophrenias | Click to show details |
| Network | Shortest path distance of core genes in the Human protein-protein interaction network | Contribution to shortest path in PPI network: 0.0226 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg24674703 | 11 | 60869960 | CD5 | 1.44E-5 | -4.592 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
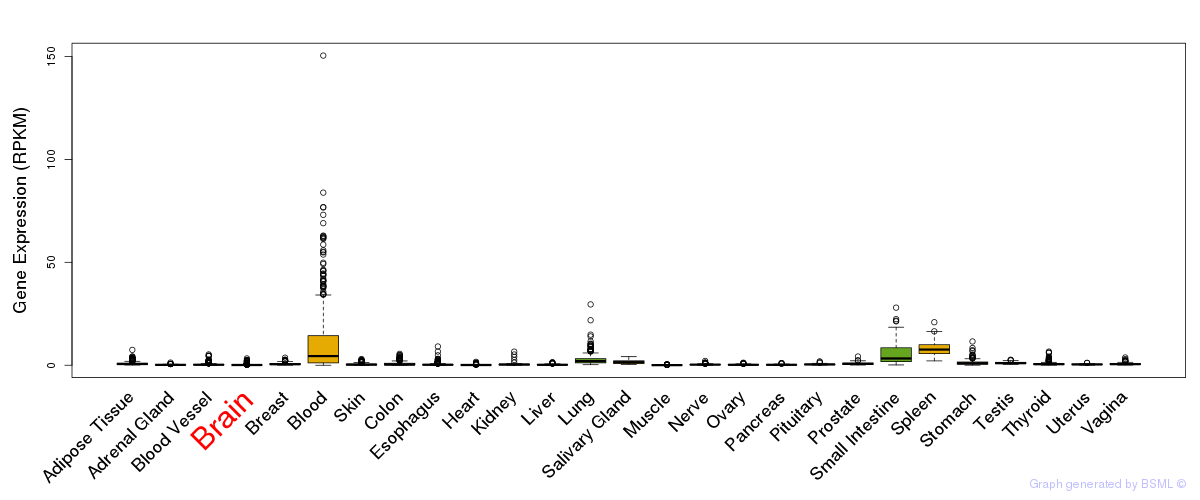
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| PODNL1 | 0.74 | 0.23 |
| KLK10 | 0.71 | 0.21 |
| EGFL6 | 0.67 | 0.15 |
| GPR177 | 0.66 | 0.04 |
| MAP4K1 | 0.66 | 0.19 |
| ANXA3 | 0.66 | 0.36 |
| OTX2 | 0.66 | 0.11 |
| HSD11B2 | 0.66 | 0.23 |
| ANO2 | 0.65 | 0.10 |
| CEACAM21 | 0.64 | 0.20 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GTDC1 | -0.31 | -0.35 |
| EMX1 | -0.30 | -0.38 |
| SATB1 | -0.28 | -0.36 |
| ARL4D | -0.28 | -0.34 |
| ARHGEF2 | -0.27 | -0.32 |
| C1orf96 | -0.27 | -0.33 |
| CRYM | -0.27 | -0.32 |
| ZNF238 | -0.27 | -0.35 |
| MEF2C | -0.26 | -0.34 |
| LMO7 | -0.26 | -0.33 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005044 | scavenger receptor activity | IEA | - | |
| GO:0005515 | protein binding | IPI | 1711157 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0008283 | cell proliferation | NAS | 3093892 | |
| GO:0008037 | cell recognition | NAS | 1711157 | |
| GO:0008624 | induction of apoptosis by extracellular signals | IEA | - | |
| GO:0031295 | T cell costimulation | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0009897 | external side of plasma membrane | IEA | - | |
| GO:0005886 | plasma membrane | IDA | 11390434 | |
| GO:0005887 | integral to plasma membrane | NAS | 1711157 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| CAMK2D | CAMKD | DKFZp686G23119 | DKFZp686I2288 | MGC44911 | calcium/calmodulin-dependent protein kinase II delta | in vitro Two-hybrid | BioGRID | 9692886 |
| CBL | C-CBL | CBL2 | RNF55 | Cas-Br-M (murine) ecotropic retroviral transforming sequence | - | HPRD,BioGRID | 9603468 |
| CD2 | SRBC | T11 | CD2 molecule | - | HPRD,BioGRID | 10510361 |
| CD247 | CD3-ZETA | CD3H | CD3Q | CD3Z | T3Z | TCRZ | CD247 molecule | - | HPRD | 1385158 |
| CD27 | MGC20393 | S152 | T14 | TNFRSF7 | Tp55 | CD27 molecule | Reconstituted Complex | BioGRID | 1711157 |
| CD4 | CD4mut | CD4 molecule | - | HPRD | 7539755 |
| CD6 | FLJ44171 | TP120 | CD6 molecule | - | HPRD,BioGRID | 12473675 |
| CD72 | CD72b | LYB2 | CD72 molecule | - | HPRD | 1711157 |8806810 |
| DYNLT3 | RP3 | TCTE1L | TCTEX1L | dynein, light chain, Tctex-type 3 | - | HPRD,BioGRID | 9692886 |
| LCK | YT16 | p56lck | pp58lck | lymphocyte-specific protein tyrosine kinase | - | HPRD | 7513045 |
| PIK3R1 | GRB1 | p85 | p85-ALPHA | phosphoinositide-3-kinase, regulatory subunit 1 (alpha) | - | HPRD,BioGRID | 9079809 |
| PTPN6 | HCP | HCPH | HPTP1C | PTP-1C | SH-PTP1 | SHP-1 | SHP-1L | SHP1 | protein tyrosine phosphatase, non-receptor type 6 | - | HPRD,BioGRID | 10082557 |
| RASA1 | CM-AVM | CMAVM | DKFZp434N071 | GAP | PKWS | RASA | RASGAP | p120GAP | p120RASGAP | RAS p21 protein activator (GTPase activating protein) 1 | - | HPRD,BioGRID | 9603468 |
| ZAP70 | FLJ17670 | FLJ17679 | SRK | STD | TZK | ZAP-70 | zeta-chain (TCR) associated protein kinase 70kDa | - | HPRD,BioGRID | 9378960 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG HEMATOPOIETIC CELL LINEAGE | 88 | 60 | All SZGR 2.0 genes in this pathway |
| BIOCARTA DC PATHWAY | 22 | 20 | All SZGR 2.0 genes in this pathway |
| HUTTMANN B CLL POOR SURVIVAL UP | 276 | 187 | All SZGR 2.0 genes in this pathway |
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| JAATINEN HEMATOPOIETIC STEM CELL DN | 226 | 132 | All SZGR 2.0 genes in this pathway |
| SHANK TAL1 TARGETS DN | 10 | 6 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 11Q12 Q14 AMPLICON | 158 | 93 | All SZGR 2.0 genes in this pathway |
| BYSTRYKH HEMATOPOIESIS STEM CELL QTL TRANS | 882 | 572 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| KUMAR TARGETS OF MLL AF9 FUSION | 405 | 264 | All SZGR 2.0 genes in this pathway |
| ZHOU TNF SIGNALING 30MIN | 54 | 36 | All SZGR 2.0 genes in this pathway |
| HILLION HMGA1B TARGETS | 92 | 68 | All SZGR 2.0 genes in this pathway |
| ACEVEDO METHYLATED IN LIVER CANCER DN | 940 | 425 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS DN | 366 | 257 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE UP | 857 | 456 | All SZGR 2.0 genes in this pathway |
| BOSCO TH1 CYTOTOXIC MODULE | 114 | 62 | All SZGR 2.0 genes in this pathway |