Gene Page: BZRAP1
Summary ?
| GeneID | 9256 |
| Symbol | BZRAP1 |
| Synonyms | PBR-IP|PRAX-1|PRAX1|RIM-BP1|RIMBP1 |
| Description | benzodiazepine receptor (peripheral) associated protein 1 |
| Reference | MIM:610764|HGNC:HGNC:16831|Ensembl:ENSG00000005379|HPRD:09832|Vega:OTTHUMG00000178923 |
| Gene type | protein-coding |
| Map location | 17q22 |
| Pascal p-value | 0.012 |
| Sherlock p-value | 0.253 |
| Fetal beta | -0.359 |
| DMG | 2 (# studies) |
| eGene | Cerebellar Hemisphere Cerebellum |
| Support | GPCR SIGNALLING CompositeSet Darnell FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:vanEijk_2014 | Genome-wide DNA methylation analysis | This dataset includes 432 differentially methylated CpG sites corresponding to 391 unique transcripts between schizophrenia patients (n=260) and unaffected controls (n=250). | 4 |
| DMG:Wockner_2014 | Genome-wide DNA methylation analysis | This dataset includes 4641 differentially methylated probes corresponding to 2929 unique genes between schizophrenia patients (n=24) and controls (n=24). | 4 |
| DNM:Fromer_2014 | Whole Exome Sequencing analysis | This study reported a WES study of 623 schizophrenia trios, reporting DNMs using genomic DNA. |
Section I. Genetics and epigenetics annotation
 DNM table
DNM table
| Gene | Chromosome | Position | Ref | Alt | Transcript | AA change | Mutation type | Sift | CG46 | Trait | Study |
|---|---|---|---|---|---|---|---|---|---|---|---|
| BZRAP1 | chr17 | 56383196 | G | C | NM_001261835 NM_001261835 NM_004758 NM_004758 NM_024418 NM_024418 | . p.1743P>R . p.1752P>R . p.1692P>R | splice missense splice missense splice missense | Schizophrenia | DNM:Fromer_2014 |
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg20783697 | 17 | 56407147 | BZRAP1 | 2.406E-4 | 0.45 | 0.037 | DMG:Wockner_2014 |
| cg22182666 | 17 | 56405155 | BZRAP1 | 5.083E-4 | 0.419 | 0.047 | DMG:Wockner_2014 |
| cg18503387 | 17 | 55927006 | BZRAP1 | 0.001 | 3.232 | DMG:vanEijk_2014 | |
| cg10728503 | 17 | 56294088 | BZRAP1 | 4.629E-4 | -4.368 | DMG:vanEijk_2014 |
Section II. Transcriptome annotation
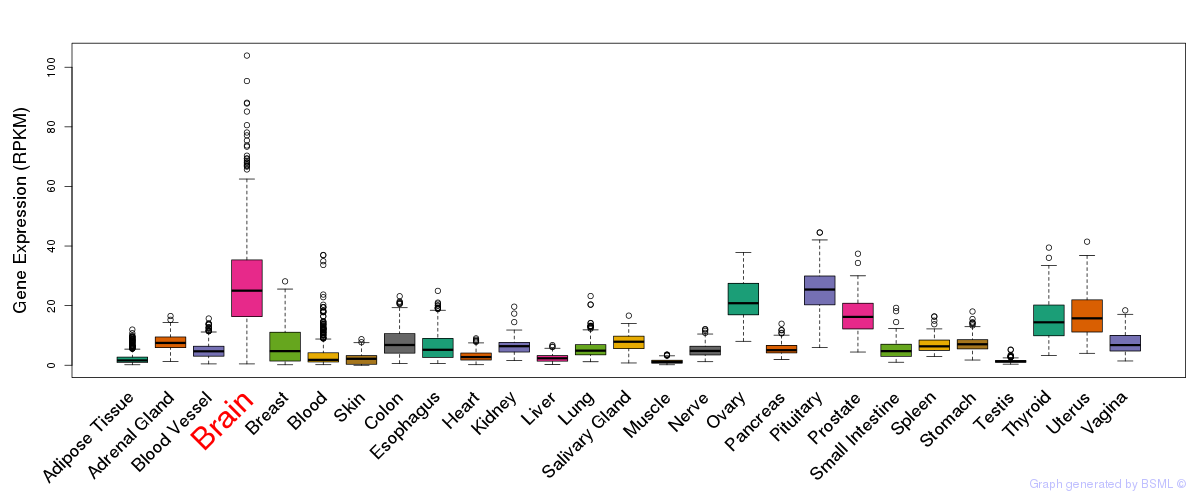
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| LRRC14 | 0.89 | 0.87 |
| POLR2C | 0.89 | 0.85 |
| GNPDA1 | 0.88 | 0.87 |
| RMND5B | 0.88 | 0.86 |
| ASB6 | 0.88 | 0.86 |
| SURF4 | 0.87 | 0.86 |
| UBL4A | 0.87 | 0.85 |
| PI4KB | 0.87 | 0.84 |
| IP6K1 | 0.87 | 0.85 |
| TBRG4 | 0.87 | 0.82 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.21 | -0.73 | -0.69 |
| AF347015.8 | -0.72 | -0.66 |
| MT-CO2 | -0.69 | -0.63 |
| AF347015.31 | -0.68 | -0.63 |
| AF347015.18 | -0.68 | -0.71 |
| MT-CYB | -0.68 | -0.63 |
| AL139819.3 | -0.67 | -0.68 |
| AF347015.27 | -0.67 | -0.65 |
| MT-ATP8 | -0.67 | -0.68 |
| AF347015.15 | -0.66 | -0.62 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| GINESTIER BREAST CANCER 20Q13 AMPLIFICATION DN | 180 | 101 | All SZGR 2.0 genes in this pathway |
| OSWALD HEMATOPOIETIC STEM CELL IN COLLAGEN GEL DN | 275 | 168 | All SZGR 2.0 genes in this pathway |
| DELYS THYROID CANCER DN | 232 | 154 | All SZGR 2.0 genes in this pathway |
| CONCANNON APOPTOSIS BY EPOXOMICIN DN | 172 | 112 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| RICKMAN TUMOR DIFFERENTIATED WELL VS POORLY UP | 236 | 139 | All SZGR 2.0 genes in this pathway |
| NIKOLSKY BREAST CANCER 17Q21 Q25 AMPLICON | 335 | 181 | All SZGR 2.0 genes in this pathway |
| HADDAD T LYMPHOCYTE AND NK PROGENITOR DN | 63 | 41 | All SZGR 2.0 genes in this pathway |
| VERHAAK AML WITH NPM1 MUTATED DN | 246 | 180 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE INCIPIENT DN | 165 | 106 | All SZGR 2.0 genes in this pathway |
| DEBIASI APOPTOSIS BY REOVIRUS INFECTION DN | 287 | 208 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| MIKKELSEN ES ICP WITH H3K4ME3 | 718 | 401 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION | 456 | 287 | All SZGR 2.0 genes in this pathway |
| MARTENS TRETINOIN RESPONSE DN | 841 | 431 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |
| TORCHIA TARGETS OF EWSR1 FLI1 FUSION UP | 271 | 165 | All SZGR 2.0 genes in this pathway |
| SERVITJA ISLET HNF1A TARGETS UP | 163 | 111 | All SZGR 2.0 genes in this pathway |
| PEDRIOLI MIR31 TARGETS DN | 418 | 245 | All SZGR 2.0 genes in this pathway |