Gene Page: ZNHIT3
Summary ?
| GeneID | 9326 |
| Symbol | ZNHIT3 |
| Synonyms | TRIP3 |
| Description | zinc finger HIT-type containing 3 |
| Reference | MIM:604500|HGNC:HGNC:12309|Ensembl:ENSG00000273611|HPRD:05139|Vega:OTTHUMG00000188436 |
| Gene type | protein-coding |
| Map location | 17q12 |
| Pascal p-value | 3.793E-4 |
| Sherlock p-value | 0.65 |
| Fetal beta | 0.344 |
| DMG | 1 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CNV:YES | Copy number variation studies | Manual curation | |
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg25975805 | 17 | 34838641 | ZNHIT3 | 4.48E-8 | -0.013 | 1.23E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
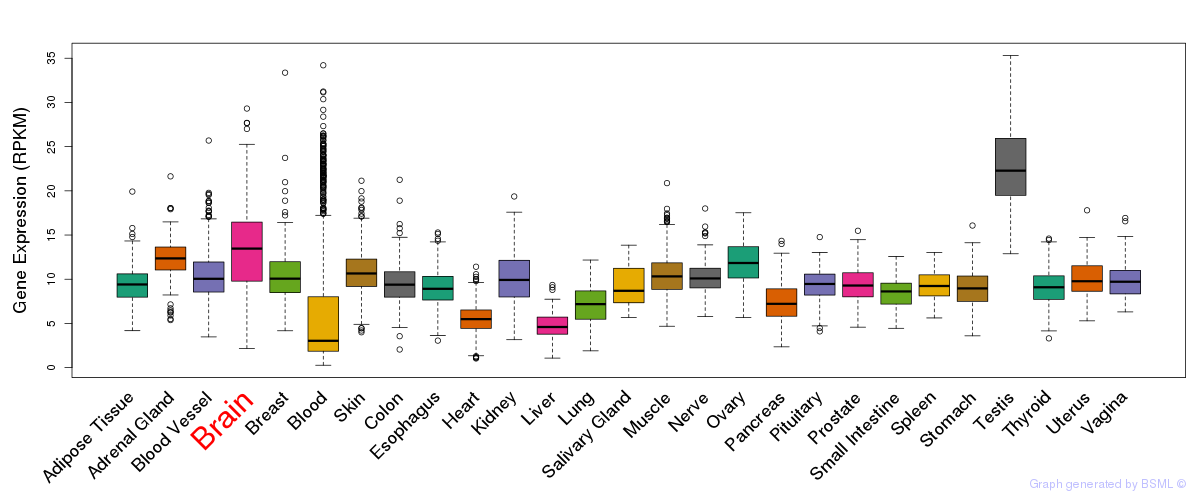
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GNAZ | 0.87 | 0.89 |
| PACS1 | 0.87 | 0.90 |
| PIK3R2 | 0.86 | 0.90 |
| SNX26 | 0.85 | 0.88 |
| GPRIN1 | 0.84 | 0.88 |
| KCTD13 | 0.84 | 0.88 |
| TNRC4 | 0.84 | 0.88 |
| MAST1 | 0.84 | 0.86 |
| MAPKBP1 | 0.84 | 0.89 |
| PCDHGC4 | 0.84 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.31 | -0.67 | -0.78 |
| AF347015.27 | -0.67 | -0.78 |
| MT-CO2 | -0.66 | -0.79 |
| MT-CYB | -0.65 | -0.76 |
| AF347015.33 | -0.64 | -0.74 |
| AF347015.8 | -0.64 | -0.77 |
| S100B | -0.63 | -0.72 |
| AF347015.15 | -0.62 | -0.74 |
| C5orf53 | -0.61 | -0.65 |
| AF347015.2 | -0.61 | -0.76 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HAHTOLA MYCOSIS FUNGOIDES SKIN UP | 177 | 113 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP | 1142 | 669 | All SZGR 2.0 genes in this pathway |
| PUJANA XPRSS INT NETWORK | 168 | 103 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA2 PCC NETWORK | 423 | 265 | All SZGR 2.0 genes in this pathway |
| PUJANA ATM PCC NETWORK | 1442 | 892 | All SZGR 2.0 genes in this pathway |
| PUJANA CHEK2 PCC NETWORK | 779 | 480 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA CENTERED NETWORK | 117 | 72 | All SZGR 2.0 genes in this pathway |
| GEORGES TARGETS OF MIR192 AND MIR215 | 893 | 528 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS DN | 366 | 238 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE DN | 1237 | 837 | All SZGR 2.0 genes in this pathway |
| MARTINEZ RESPONSE TO TRABECTEDIN DN | 271 | 175 | All SZGR 2.0 genes in this pathway |
| ALONSO METASTASIS UP | 198 | 128 | All SZGR 2.0 genes in this pathway |
| SWEET LUNG CANCER KRAS UP | 491 | 316 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS CALB1 CORR UP | 548 | 370 | All SZGR 2.0 genes in this pathway |
| FORTSCHEGGER PHF8 TARGETS DN | 784 | 464 | All SZGR 2.0 genes in this pathway |
| HOLLEMAN ASPARAGINASE RESISTANCE ALL DN | 25 | 14 | All SZGR 2.0 genes in this pathway |