Gene Page: PPT2
Summary ?
| GeneID | 9374 |
| Symbol | PPT2 |
| Synonyms | C6orf8|G14|PPT-2 |
| Description | palmitoyl-protein thioesterase 2 |
| Reference | MIM:603298|HGNC:HGNC:9326|Ensembl:ENSG00000221988|HPRD:04486|Vega:OTTHUMG00000031257 |
| Gene type | protein-coding |
| Map location | 6p21.3 |
| Pascal p-value | 1E-12 |
| Sherlock p-value | 0.853 |
| Fetal beta | -0.253 |
| DMG | 2 (# studies) |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 2 |
| DMG:Montano_2016 | Genome-wide DNA methylation analysis | This dataset includes 172 replicated associations between CpGs with schizophrenia. | 2 |
| GSMA_I | Genome scan meta-analysis | Psr: 0.033 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg06902929 | 6 | 32123258 | PPT2 | 1.27E-4 | -0.004 | 0.134 | DMG:Montano_2016 |
| cg09672152 | 6 | 32122102 | PPT2 | 1.15E-8 | -0.015 | 4.8E-6 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
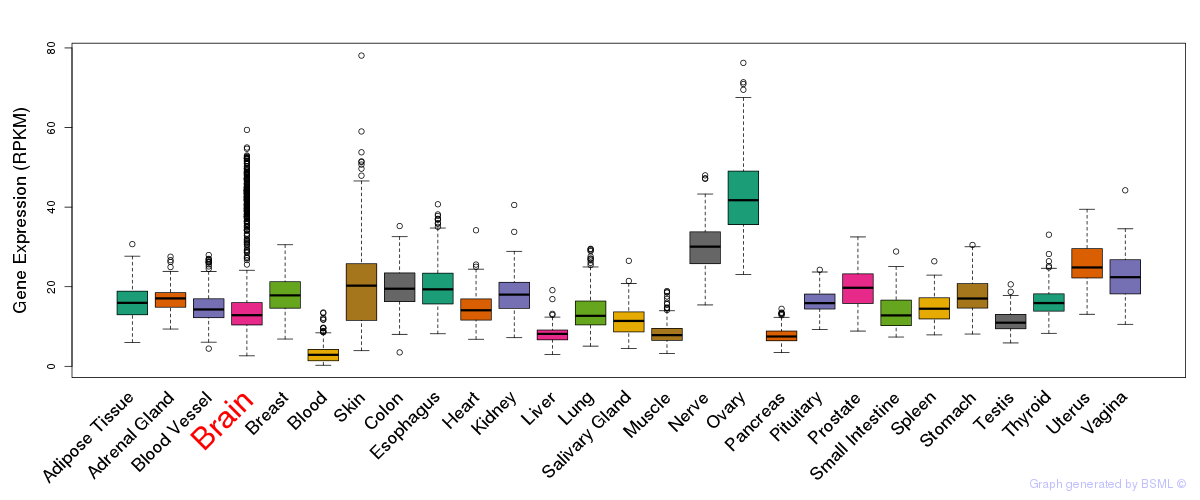
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| OXR1 | 0.93 | 0.89 |
| ARHGAP26 | 0.93 | 0.85 |
| SLC4A10 | 0.93 | 0.88 |
| EFR3A | 0.93 | 0.84 |
| NCOA7 | 0.92 | 0.87 |
| HTR2A | 0.92 | 0.85 |
| SYNJ1 | 0.91 | 0.88 |
| GLS | 0.91 | 0.86 |
| ITPR1 | 0.91 | 0.81 |
| ARFGEF2 | 0.91 | 0.84 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.48 | -0.61 |
| AC006276.2 | -0.44 | -0.44 |
| TBC1D10A | -0.44 | -0.37 |
| FADS2 | -0.41 | -0.28 |
| EFEMP2 | -0.40 | -0.40 |
| RPLP1 | -0.40 | -0.44 |
| NME4 | -0.40 | -0.52 |
| HEBP2 | -0.39 | -0.52 |
| SLC38A5 | -0.39 | -0.45 |
| RPL23A | -0.39 | -0.42 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0016787 | hydrolase activity | IEA | - | |
| GO:0008474 | palmitoyl-(protein) hydrolase activity | NAS | 10417332 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006464 | protein modification process | NAS | 9341199 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005764 | lysosome | NAS | 9341199 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG LYSOSOME | 121 | 83 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS UP | 137 | 94 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| BOGNI TREATMENT RELATED MYELOID LEUKEMIA DN | 33 | 19 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 1 DN | 242 | 165 | All SZGR 2.0 genes in this pathway |
| MISSIAGLIA REGULATED BY METHYLATION UP | 126 | 78 | All SZGR 2.0 genes in this pathway |
| PATIL LIVER CANCER | 747 | 453 | All SZGR 2.0 genes in this pathway |
| NUYTTEN NIPP1 TARGETS UP | 769 | 437 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP | 1037 | 673 | All SZGR 2.0 genes in this pathway |
| MANTOVANI VIRAL GPCR SIGNALING DN | 49 | 37 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| MAHAJAN RESPONSE TO IL1A DN | 76 | 57 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 48HR UP | 180 | 125 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| LIU VAV3 PROSTATE CARCINOGENESIS DN | 17 | 14 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 4 UP | 112 | 64 | All SZGR 2.0 genes in this pathway |
| FIGUEROA AML METHYLATION CLUSTER 6 UP | 140 | 81 | All SZGR 2.0 genes in this pathway |
| PANGAS TUMOR SUPPRESSION BY SMAD1 AND SMAD5 DN | 157 | 106 | All SZGR 2.0 genes in this pathway |
| LINSLEY MIR16 TARGETS | 206 | 127 | All SZGR 2.0 genes in this pathway |
| KOINUMA TARGETS OF SMAD2 OR SMAD3 | 824 | 528 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-141/200a | 541 | 547 | m8 | hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG |
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| hsa-miR-141 | UAACACUGUCUGGUAAAGAUGG | ||||
| hsa-miR-200a | UAACACUGUCUGGUAACGAUGU | ||||
| miR-143 | 574 | 580 | 1A | hsa-miR-143brain | UGAGAUGAAGCACUGUAGCUCA |
| miR-326 | 35 | 41 | 1A | hsa-miR-326 | CCUCUGGGCCCUUCCUCCAG |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.