Gene Page: TM9SF2
Summary ?
| GeneID | 9375 |
| Symbol | TM9SF2 |
| Synonyms | P76 |
| Description | transmembrane 9 superfamily member 2 |
| Reference | MIM:604678|HGNC:HGNC:11865|Ensembl:ENSG00000125304|HPRD:09198|Vega:OTTHUMG00000017272 |
| Gene type | protein-coding |
| Map location | 13q32.3 |
| Pascal p-value | 0.722 |
| Sherlock p-value | 0.128 |
| Fetal beta | 0.967 |
| DMG | 1 (# studies) |
| eGene | Cerebellar Hemisphere |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg26455579 | 13 | 100153786 | TM9SF2 | 3.41E-8 | -0.005 | 1.01E-5 | DMG:Jaffe_2016 |
Section II. Transcriptome annotation
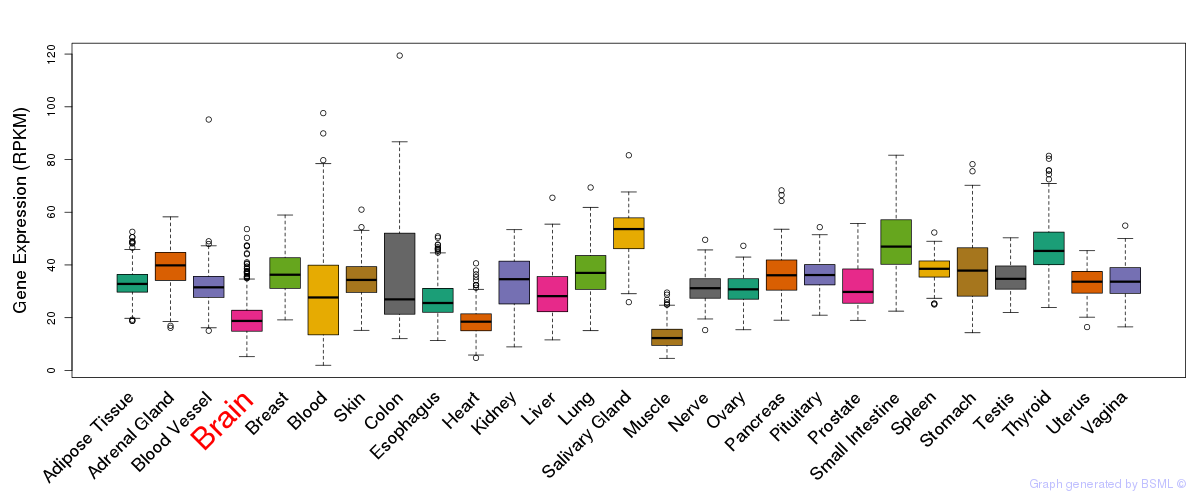
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| GAB1 | 0.81 | 0.83 |
| TMTC2 | 0.81 | 0.84 |
| M6PRBP1 | 0.80 | 0.82 |
| MEGF10 | 0.80 | 0.82 |
| RFTN2 | 0.79 | 0.83 |
| LASS2 | 0.79 | 0.84 |
| CA14 | 0.78 | 0.77 |
| MAGT1 | 0.78 | 0.82 |
| COL4A5 | 0.78 | 0.82 |
| PRR5L | 0.78 | 0.83 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| KLHL1 | -0.39 | -0.25 |
| MPPED1 | -0.34 | -0.25 |
| NEUROD6 | -0.34 | -0.28 |
| DACT1 | -0.34 | -0.20 |
| SATB2 | -0.33 | -0.22 |
| RP9P | -0.33 | -0.44 |
| AC011491.1 | -0.33 | -0.38 |
| IER5L | -0.33 | -0.24 |
| MEF2C | -0.32 | -0.25 |
| RPRM | -0.32 | -0.27 |
Section III. Gene Ontology annotation
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0006810 | transport | TAS | 9729438 | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005768 | endosome | TAS | 9729438 | |
| GO:0016020 | membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9729438 | |
| GO:0010008 | endosome membrane | IEA | - |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| WANG LMO4 TARGETS UP | 372 | 227 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE DN | 485 | 334 | All SZGR 2.0 genes in this pathway |
| OLSSON E2F3 TARGETS UP | 28 | 14 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| TOMLINS METASTASIS DN | 20 | 16 | All SZGR 2.0 genes in this pathway |
| SPIELMAN LYMPHOBLAST EUROPEAN VS ASIAN DN | 584 | 395 | All SZGR 2.0 genes in this pathway |
| TARTE PLASMA CELL VS B LYMPHOCYTE UP | 78 | 51 | All SZGR 2.0 genes in this pathway |
| CHEOK RESPONSE TO MERCAPTOPURINE AND LD MTX UP | 10 | 6 | All SZGR 2.0 genes in this pathway |
| WELCSH BRCA1 TARGETS UP | 198 | 132 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA CHEMOTAXIS DN | 457 | 302 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN | 668 | 419 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS UNANNOTATED DN | 193 | 112 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN | 553 | 343 | All SZGR 2.0 genes in this pathway |
| PECE MAMMARY STEM CELL DN | 146 | 88 | All SZGR 2.0 genes in this pathway |