Gene Page: MED23
Summary ?
| GeneID | 9439 |
| Symbol | MED23 |
| Synonyms | ARC130|CRSP130|CRSP133|CRSP3|DRIP130|MRT18|SUR-2|SUR2 |
| Description | mediator complex subunit 23 |
| Reference | MIM:605042|HGNC:HGNC:2372|Ensembl:ENSG00000112282|HPRD:05436|Vega:OTTHUMG00000015565 |
| Gene type | protein-coding |
| Map location | 6q22.33-q24.1 |
| Pascal p-value | 0.101 |
| Fetal beta | 0.651 |
| DMG | 1 (# studies) |
| eGene | Myers' cis & trans |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg05527918 | 6 | 131949098 | MED23 | 4.06E-9 | -0.012 | 2.49E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs11120206 | chr1 | 206610593 | MED23 | 9439 | 0.03 | trans | ||
| rs2716734 | chr2 | 39947720 | MED23 | 9439 | 0.14 | trans | ||
| rs2716736 | chr2 | 39947946 | MED23 | 9439 | 0.14 | trans | ||
| rs11754465 | chr6 | 11761260 | MED23 | 9439 | 0.01 | trans | ||
| rs16893142 | chr8 | 127104392 | MED23 | 9439 | 0.19 | trans | ||
| rs12255258 | chr10 | 106306927 | MED23 | 9439 | 8.841E-4 | trans | ||
| rs6128541 | chr20 | 57917688 | MED23 | 9439 | 0.08 | trans |
Section II. Transcriptome annotation
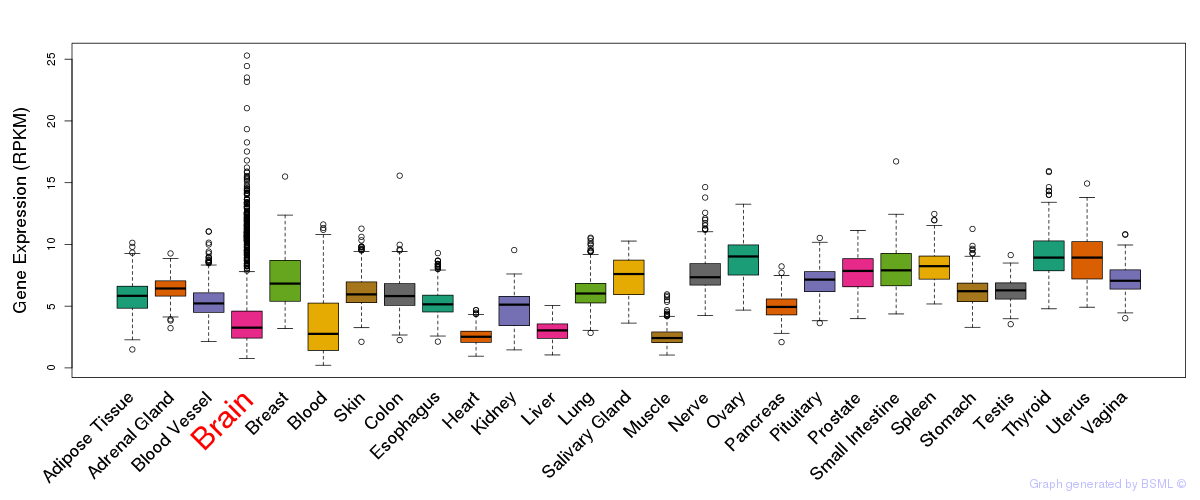
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| SYNJ1 | 0.95 | 0.93 |
| GABRG2 | 0.95 | 0.93 |
| NCOA7 | 0.95 | 0.92 |
| GLS | 0.95 | 0.93 |
| ATP6V1A | 0.94 | 0.92 |
| FAM179B | 0.94 | 0.88 |
| PIK3CB | 0.93 | 0.94 |
| NSF | 0.93 | 0.90 |
| ARFGEF2 | 0.93 | 0.89 |
| MAP2K4 | 0.93 | 0.92 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| EFEMP2 | -0.47 | -0.53 |
| RAB34 | -0.47 | -0.57 |
| C1orf61 | -0.44 | -0.59 |
| IRF7 | -0.43 | -0.56 |
| GATSL3 | -0.42 | -0.46 |
| AL022328.1 | -0.42 | -0.53 |
| DBI | -0.41 | -0.47 |
| AC006276.2 | -0.41 | -0.41 |
| CLEC3B | -0.41 | -0.52 |
| RAB13 | -0.41 | -0.50 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0003713 | transcription coactivator activity | TAS | 9989412 | |
| GO:0005515 | protein binding | IPI | 17438371 | |
| GO:0030528 | transcription regulator activity | IDA | 9989412 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0006367 | transcription initiation from RNA polymerase II promoter | TAS | 9989412 | |
| GO:0006357 | regulation of transcription from RNA polymerase II promoter | TAS | 9989412 | |
| GO:0006350 | transcription | IEA | - | |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0005634 | nucleus | IEA | - | |
| GO:0005667 | transcription factor complex | IDA | 9989412 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| BRCA1 | BRCAI | BRCC1 | IRIS | PSCP | RNF53 | breast cancer 1, early onset | Affinity Capture-MS | BioGRID | 15208681 |
| CDK8 | K35 | MGC126074 | MGC126075 | cyclin-dependent kinase 8 | Affinity Capture-Western | BioGRID | 11416138 |
| CEBPB | C/EBP-beta | CRP2 | IL6DBP | LAP | MGC32080 | NF-IL6 | TCF5 | CCAAT/enhancer binding protein (C/EBP), beta | - | HPRD,BioGRID | 14759369 |
| ELF3 | EPR-1 | ERT | ESE-1 | ESX | E74-like factor 3 (ets domain transcription factor, epithelial-specific ) | - | HPRD,BioGRID | 12242338 |
| FOXJ3 | MGC165036 | MGC176686 | forkhead box J3 | - | HPRD | 12421765 |
| HNF4A | FLJ39654 | HNF4 | HNF4a7 | HNF4a8 | HNF4a9 | MODY | MODY1 | NR2A1 | NR2A21 | TCF | TCF14 | hepatocyte nuclear factor 4, alpha | Affinity Capture-Western | BioGRID | 12089346 |
| IRF1 | IRF-1 | MAR | interferon regulatory factor 1 | An unspecified isoform of MED23 interacts with IRF-1 promoter. | BIND | 15893730 |
| MED10 | L6 | MGC5309 | NUT2 | TRG20 | mediator complex subunit 10 | Affinity Capture-MS | BioGRID | 15175163 |
| MED19 | DT2P1G7 | LCMR1 | mediator complex subunit 19 | Affinity Capture-MS | BioGRID | 15175163 |
| MED26 | CRSP7 | CRSP70 | mediator complex subunit 26 | Affinity Capture-MS | BioGRID | 15175163 |
| MED28 | 1500003D12Rik | DKFZp434N185 | EG1 | magicin | mediator complex subunit 28 | Affinity Capture-MS | BioGRID | 15175163 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | - | HPRD | 14576168 |
| MED29 | DKFZp434H247 | IXL | mediator complex subunit 29 | Affinity Capture-MS | BioGRID | 15175163 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | - | HPRD | 14638676 |
| MED9 | FLJ10193 | MED25 | MGC138234 | mediator complex subunit 9 | Affinity Capture-MS | BioGRID | 15175163 |
| NCOA6 | AIB3 | ANTP | ASC2 | HOX1.1 | HOXA7 | KIAA0181 | NRC | PRIP | RAP250 | TRBP | nuclear receptor coactivator 6 | - | HPRD,BioGRID | 10823961 |
| WDTC1 | ADP | KIAA1037 | RP11-4K3__A.1 | WD and tetratricopeptide repeats 1 | - | HPRD | 12421765 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| REACTOME DEVELOPMENTAL BIOLOGY | 396 | 292 | All SZGR 2.0 genes in this pathway |
| REACTOME PPARA ACTIVATES GENE EXPRESSION | 104 | 72 | All SZGR 2.0 genes in this pathway |
| REACTOME GENERIC TRANSCRIPTION PATHWAY | 352 | 181 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS | 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME FATTY ACID TRIACYLGLYCEROL AND KETONE BODY METABOLISM | 168 | 115 | All SZGR 2.0 genes in this pathway |
| REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | 72 | 53 | All SZGR 2.0 genes in this pathway |
| RODRIGUES THYROID CARCINOMA POORLY DIFFERENTIATED DN | 805 | 505 | All SZGR 2.0 genes in this pathway |
| PEREZ TP53 TARGETS | 1174 | 695 | All SZGR 2.0 genes in this pathway |
| BENPORATH NANOG TARGETS | 988 | 594 | All SZGR 2.0 genes in this pathway |
| RAMALHO STEMNESS UP | 206 | 118 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| MA PITUITARY FETAL VS ADULT DN | 19 | 17 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 3 | 720 | 440 | All SZGR 2.0 genes in this pathway |
| STEARMAN LUNG CANCER EARLY VS LATE UP | 125 | 89 | All SZGR 2.0 genes in this pathway |
| RAY TUMORIGENESIS BY ERBB2 CDC25A UP | 104 | 57 | All SZGR 2.0 genes in this pathway |
| PILON KLF1 TARGETS DN | 1972 | 1213 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE GENES | 39 | 20 | All SZGR 2.0 genes in this pathway |