Gene Page: HOMER1
Summary ?
| GeneID | 9456 |
| Symbol | HOMER1 |
| Synonyms | HOMER|HOMER1A|HOMER1B|HOMER1C|SYN47|Ves-1 |
| Description | homer scaffolding protein 1 |
| Reference | MIM:604798|HGNC:HGNC:17512|HPRD:09211| |
| Gene type | protein-coding |
| Map location | 5q14.2 |
| Pascal p-value | 0.027 |
| Sherlock p-value | 0.225 |
| Fetal beta | -1.287 |
| eGene | Hippocampus |
| Support | INTRACELLULAR SIGNAL TRANSDUCTION G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-CONSENSUS G2Cdb.human_BAYES-COLLINS-HUMAN-PSD-FULL G2Cdb.human_BAYES-COLLINS-MOUSE-PSD-CONSENSUS G2Cdb.human_mGluR5 G2Cdb.humanNRC G2Cdb.humanPSD G2Cdb.humanPSP CompositeSet Potential synaptic genes |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
| GO_Annotation | Mapping neuro-related keywords to Gene Ontology annotations | Hits with neuro-related keywords: 3 |
Section I. Genetics and epigenetics annotation
Section II. Transcriptome annotation
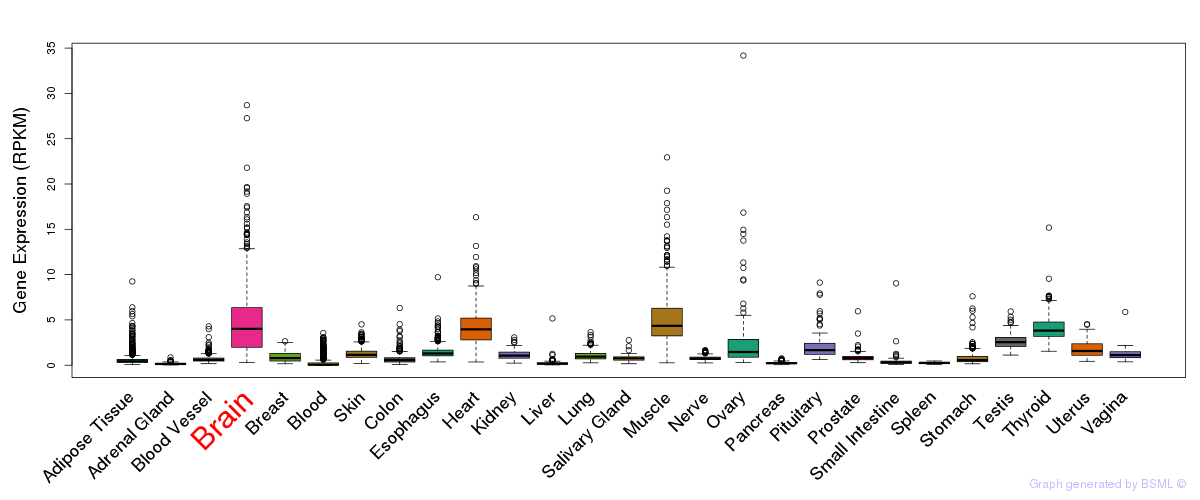
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| HOMER2 | 0.85 | 0.73 |
| REEP2 | 0.84 | 0.88 |
| GALNT14 | 0.83 | 0.66 |
| ATP13A2 | 0.83 | 0.88 |
| PPM1J | 0.82 | 0.74 |
| GRIN2D | 0.82 | 0.81 |
| PLA2G15 | 0.82 | 0.82 |
| CPNE7 | 0.81 | 0.72 |
| CHAC1 | 0.80 | 0.87 |
| CHRNA4 | 0.80 | 0.79 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| AF347015.2 | -0.41 | -0.45 |
| AF347015.31 | -0.38 | -0.44 |
| AF347015.21 | -0.38 | -0.44 |
| AF347015.8 | -0.38 | -0.44 |
| AF347015.33 | -0.37 | -0.43 |
| MT-CO2 | -0.36 | -0.44 |
| OAF | -0.35 | -0.41 |
| MT-ATP8 | -0.35 | -0.44 |
| AL139819.3 | -0.35 | -0.46 |
| AF347015.27 | -0.35 | -0.44 |
Section III. Gene Ontology annotation
| Molecular function | GO term | Evidence | Neuro keywords | PubMed ID |
|---|---|---|---|---|
| GO:0005515 | protein binding | TAS | 9808458 | |
| Biological process | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0007268 | synaptic transmission | TAS | neuron, Synap, Neurotransmitter (GO term level: 6) | 9808458 |
| GO:0007206 | metabotropic glutamate receptor, phospholipase C activating pathway | TAS | glutamate (GO term level: 10) | 9808459 |
| Cellular component | GO term | Evidence | Neuro keywords | PubMed ID |
| GO:0045211 | postsynaptic membrane | IEA | Synap, Neurotransmitter (GO term level: 5) | - |
| GO:0045202 | synapse | IEA | neuron, Synap, Neurotransmitter, Glial (GO term level: 2) | - |
| GO:0005737 | cytoplasm | IEA | - | |
| GO:0005886 | plasma membrane | IEA | - | |
| GO:0005887 | integral to plasma membrane | TAS | 9808459 | |
| GO:0030054 | cell junction | IEA | - |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| ABI3 | NESH | SSH3BP3 | ABI family, member 3 | Two-hybrid | BioGRID | 16189514 |
| AGAP2 | CENTG1 | FLJ16430 | GGAP2 | KIAA0167 | PIKE | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 | - | HPRD,BioGRID | 14528310 |
| C1orf116 | DKFZp666H2010 | FLJ36507 | MGC2742 | MGC4309 | SARG | chromosome 1 open reading frame 116 | Two-hybrid | BioGRID | 16189514 |
| DNM3 | KIAA0820 | MGC70433 | dynamin 3 | in vitro | BioGRID | 9808459 |
| GRIA1 | GLUH1 | GLUR1 | GLURA | HBGR1 | MGC133252 | glutamate receptor, ionotropic, AMPA 1 | - | HPRD,BioGRID | 9069287 |9808458 |11418862 |
| GRIK1 | EAA3 | EEA3 | GLR5 | GLUR5 | glutamate receptor, ionotropic, kainate 1 | - | HPRD,BioGRID | 9069287 |9808458 |11418862 |
| GRM1 | GPRC1A | GRM1A | MGLUR1 | MGLUR1A | mGlu1 | glutamate receptor, metabotropic 1 | - | HPRD | 9069287 |
| HOMER1 | HOMER | HOMER1A | HOMER1B | HOMER1C | SYN47 | Ves-1 | homer homolog 1 (Drosophila) | - | HPRD | 9808458 |
| HOMER3 | HOMER-3 | homer homolog 3 (Drosophila) | - | HPRD | 9808458 |
| ITPR1 | INSP3R1 | IP3R | IP3R1 | SCA15 | SCA16 | inositol 1,4,5-triphosphate receptor, type 1 | - | HPRD,BioGRID | 9808459 |
| PARK2 | AR-JP | LPRS2 | PDJ | PRKN | Parkinson disease (autosomal recessive, juvenile) 2, parkin | Affinity Capture-Western | BioGRID | 11679592 |
| RYR1 | CCO | MHS | MHS1 | RYDR | RYR | SKRR | ryanodine receptor 1 (skeletal) | - | HPRD,BioGRID | 12223488 |12810060 |14660561 |
| RYR2 | ARVC2 | ARVD2 | VTSIP | ryanodine receptor 2 (cardiac) | - | HPRD,BioGRID | 12887973 |
| TANC1 | KIAA1728 | ROLSB | TANC | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 1 | - | HPRD | 15673434 |
| TRPC1 | HTRP-1 | MGC133334 | MGC133335 | TRP1 | transient receptor potential cation channel, subfamily C, member 1 | Affinity Capture-Western | BioGRID | 14505576 |
| TRPC2 | - | transient receptor potential cation channel, subfamily C, member 2 (pseudogene) | Affinity Capture-Western | BioGRID | 14505576 |
| TRPC5 | TRP5 | transient receptor potential cation channel, subfamily C, member 5 | Affinity Capture-Western | BioGRID | 14505576 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| WINTER HYPOXIA UP | 92 | 57 | All SZGR 2.0 genes in this pathway |
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| TONKS TARGETS OF RUNX1 RUNX1T1 FUSION MONOCYTE UP | 204 | 140 | All SZGR 2.0 genes in this pathway |
| NAGASHIMA NRG1 SIGNALING UP | 176 | 123 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN | 508 | 354 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY SERUM DEPRIVATION DN | 234 | 147 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND SERUM DEPRIVATION DN | 84 | 54 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 DN | 855 | 609 | All SZGR 2.0 genes in this pathway |
| DACOSTA UV RESPONSE VIA ERCC3 COMMON DN | 483 | 336 | All SZGR 2.0 genes in this pathway |
| MOHANKUMAR TLX1 TARGETS UP | 414 | 287 | All SZGR 2.0 genes in this pathway |
| GROSS HYPOXIA VIA ELK3 AND HIF1A UP | 142 | 104 | All SZGR 2.0 genes in this pathway |
| FERREIRA EWINGS SARCOMA UNSTABLE VS STABLE UP | 167 | 92 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX | 795 | 478 | All SZGR 2.0 genes in this pathway |
| BENPORATH ES 1 | 379 | 235 | All SZGR 2.0 genes in this pathway |
| ONDER CDH1 TARGETS 2 DN | 464 | 276 | All SZGR 2.0 genes in this pathway |
| CUI TCF21 TARGETS 2 DN | 830 | 547 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 12HR UP | 111 | 68 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP | 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| SANSOM APC MYC TARGETS | 217 | 138 | All SZGR 2.0 genes in this pathway |
| SANSOM APC TARGETS REQUIRE MYC | 210 | 123 | All SZGR 2.0 genes in this pathway |
| FOSTER TOLERANT MACROPHAGE DN | 409 | 268 | All SZGR 2.0 genes in this pathway |
| HUANG FOXA2 TARGETS UP | 45 | 28 | All SZGR 2.0 genes in this pathway |
| ACEVEDO LIVER CANCER WITH H3K27ME3 UP | 295 | 149 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| WEST ADRENOCORTICAL TUMOR UP | 294 | 199 | All SZGR 2.0 genes in this pathway |
| YOSHIMURA MAPK8 TARGETS UP | 1305 | 895 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS CTNNB1 DN | 170 | 105 | All SZGR 2.0 genes in this pathway |
| CHIANG LIVER CANCER SUBCLASS POLYSOMY7 DN | 24 | 17 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG REGULATED BY MYC UP | 72 | 53 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 1HR DN | 222 | 147 | All SZGR 2.0 genes in this pathway |
| QI HYPOXIA | 140 | 96 | All SZGR 2.0 genes in this pathway |
| GOBERT OLIGODENDROCYTE DIFFERENTIATION DN | 1080 | 713 | All SZGR 2.0 genes in this pathway |
| ZWANG CLASS 1 TRANSIENTLY INDUCED BY EGF | 516 | 308 | All SZGR 2.0 genes in this pathway |
Section VI. microRNA annotation
| miRNA family | Target position | miRNA ID | miRNA seq | ||
|---|---|---|---|---|---|
| UTR start | UTR end | Match method | |||
| miR-369-3p | 1762 | 1768 | 1A | hsa-miR-369-3p | AAUAAUACAUGGUUGAUCUUU |
| miR-374 | 1762 | 1768 | m8 | hsa-miR-374 | UUAUAAUACAACCUGAUAAGUG |
| miR-495 | 1629 | 1635 | m8 | hsa-miR-495brain | AAACAAACAUGGUGCACUUCUUU |
- SZ: miRNAs which differentially expressed in brain cortex of schizophrenia patients comparing with control samples using microarray. Click here to see the list of SZ related miRNAs.
- Brain: miRNAs which are expressed in brain based on miRNA microarray expression studies. Click here to see the list of brain related miRNAs.