Gene Page: EIF4E2
Summary ?
| GeneID | 9470 |
| Symbol | EIF4E2 |
| Synonyms | 4E-LP|4EHP|EIF4EL3|IF4e |
| Description | eukaryotic translation initiation factor 4E family member 2 |
| Reference | MIM:605895|HGNC:HGNC:3293|Ensembl:ENSG00000135930|HPRD:05798|Vega:OTTHUMG00000133256 |
| Gene type | protein-coding |
| Map location | 2q37.1 |
| Pascal p-value | 0.049 |
| Sherlock p-value | 0.467 |
| Fetal beta | 0.572 |
| DMG | 1 (# studies) |
| eGene | Caudate basal ganglia Cortex Putamen basal ganglia |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| DMG:Jaffe_2016 | Genome-wide DNA methylation analysis | This dataset includes 2,104 probes/CpGs associated with SZ patients (n=108) compared to 136 controls at Bonferroni-adjusted P < 0.05. | 1 |
Section I. Genetics and epigenetics annotation
 Differentially methylated gene
Differentially methylated gene
| Probe | Chromosome | Position | Nearest gene | P (dis) | Beta (dis) | FDR (dis) | Study |
|---|---|---|---|---|---|---|---|
| cg15779458 | 2 | 233415951 | EIF4E2 | 1.07E-8 | -0.024 | 4.57E-6 | DMG:Jaffe_2016 |
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs1190453 | 2 | 233417312 | EIF4E2 | ENSG00000135930.9 | 7.725E-7 | 0.01 | 1796 | gtex_brain_putamen_basal |
| rs34854862 | 2 | 233417394 | EIF4E2 | ENSG00000135930.9 | 2.829E-6 | 0.01 | 1878 | gtex_brain_putamen_basal |
| rs1190452 | 2 | 233417552 | EIF4E2 | ENSG00000135930.9 | 1.025E-6 | 0.01 | 2036 | gtex_brain_putamen_basal |
| rs377195442 | 2 | 233417932 | EIF4E2 | ENSG00000135930.9 | 1.276E-6 | 0.01 | 2416 | gtex_brain_putamen_basal |
| rs1190451 | 2 | 233418288 | EIF4E2 | ENSG00000135930.9 | 1.746E-6 | 0.01 | 2772 | gtex_brain_putamen_basal |
| rs1190450 | 2 | 233418302 | EIF4E2 | ENSG00000135930.9 | 1.745E-6 | 0.01 | 2786 | gtex_brain_putamen_basal |
| rs11381056 | 2 | 233418578 | EIF4E2 | ENSG00000135930.9 | 5.455E-7 | 0.01 | 3062 | gtex_brain_putamen_basal |
| rs1190448 | 2 | 233419084 | EIF4E2 | ENSG00000135930.9 | 1.63E-6 | 0.01 | 3568 | gtex_brain_putamen_basal |
| rs1201451 | 2 | 233419396 | EIF4E2 | ENSG00000135930.9 | 1.782E-6 | 0.01 | 3880 | gtex_brain_putamen_basal |
| rs1307667 | 2 | 233419972 | EIF4E2 | ENSG00000135930.9 | 1.52E-6 | 0.01 | 4456 | gtex_brain_putamen_basal |
| rs1190447 | 2 | 233420052 | EIF4E2 | ENSG00000135930.9 | 1.782E-6 | 0.01 | 4536 | gtex_brain_putamen_basal |
| rs201284540 | 2 | 233420652 | EIF4E2 | ENSG00000135930.9 | 1.782E-6 | 0.01 | 5136 | gtex_brain_putamen_basal |
| rs1190445 | 2 | 233420827 | EIF4E2 | ENSG00000135930.9 | 1.516E-6 | 0.01 | 5311 | gtex_brain_putamen_basal |
| rs397740637 | 2 | 233421284 | EIF4E2 | ENSG00000135930.9 | 1.782E-6 | 0.01 | 5768 | gtex_brain_putamen_basal |
| rs1729245 | 2 | 233424891 | EIF4E2 | ENSG00000135930.9 | 1.782E-6 | 0.01 | 9375 | gtex_brain_putamen_basal |
| rs1550096 | 2 | 233428385 | EIF4E2 | ENSG00000135930.9 | 1.782E-6 | 0.01 | 12869 | gtex_brain_putamen_basal |
| rs13394993 | 2 | 233437268 | EIF4E2 | ENSG00000135930.9 | 1.782E-6 | 0.01 | 21752 | gtex_brain_putamen_basal |
| rs2293338 | 2 | 233438786 | EIF4E2 | ENSG00000135930.9 | 1.782E-6 | 0.01 | 23270 | gtex_brain_putamen_basal |
| rs10185980 | 2 | 233439851 | EIF4E2 | ENSG00000135930.9 | 1.775E-6 | 0.01 | 24335 | gtex_brain_putamen_basal |
| rs12617363 | 2 | 233440296 | EIF4E2 | ENSG00000135930.9 | 7.74E-7 | 0.01 | 24780 | gtex_brain_putamen_basal |
| rs62191571 | 2 | 233440756 | EIF4E2 | ENSG00000135930.9 | 8.996E-7 | 0.01 | 25240 | gtex_brain_putamen_basal |
| rs7571154 | 2 | 233440924 | EIF4E2 | ENSG00000135930.9 | 7.24E-7 | 0.01 | 25408 | gtex_brain_putamen_basal |
| rs7571716 | 2 | 233441420 | EIF4E2 | ENSG00000135930.9 | 7.781E-7 | 0.01 | 25904 | gtex_brain_putamen_basal |
| rs13393692 | 2 | 233442016 | EIF4E2 | ENSG00000135930.9 | 1.07E-6 | 0.01 | 26500 | gtex_brain_putamen_basal |
| rs13393800 | 2 | 233442091 | EIF4E2 | ENSG00000135930.9 | 7.735E-7 | 0.01 | 26575 | gtex_brain_putamen_basal |
| rs3841083 | 2 | 233446259 | EIF4E2 | ENSG00000135930.9 | 7.587E-7 | 0.01 | 30743 | gtex_brain_putamen_basal |
| rs72243817 | 2 | 233448850 | EIF4E2 | ENSG00000135930.9 | 7.883E-7 | 0.01 | 33334 | gtex_brain_putamen_basal |
| rs12620925 | 2 | 233450820 | EIF4E2 | ENSG00000135930.9 | 1.72E-7 | 0.01 | 35304 | gtex_brain_putamen_basal |
| rs12617649 | 2 | 233450824 | EIF4E2 | ENSG00000135930.9 | 1.72E-7 | 0.01 | 35308 | gtex_brain_putamen_basal |
| rs2276559 | 2 | 233451163 | EIF4E2 | ENSG00000135930.9 | 4.718E-7 | 0.01 | 35647 | gtex_brain_putamen_basal |
| rs60029373 | 2 | 233456262 | EIF4E2 | ENSG00000135930.9 | 6.042E-7 | 0.01 | 40746 | gtex_brain_putamen_basal |
| rs4468805 | 2 | 233459663 | EIF4E2 | ENSG00000135930.9 | 1.601E-7 | 0.01 | 44147 | gtex_brain_putamen_basal |
| rs4555341 | 2 | 233459738 | EIF4E2 | ENSG00000135930.9 | 1.643E-7 | 0.01 | 44222 | gtex_brain_putamen_basal |
| rs10193553 | 2 | 233459862 | EIF4E2 | ENSG00000135930.9 | 1.739E-7 | 0.01 | 44346 | gtex_brain_putamen_basal |
| rs9808024 | 2 | 233462273 | EIF4E2 | ENSG00000135930.9 | 3.161E-6 | 0.01 | 46757 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
General gene expression (GTEx)
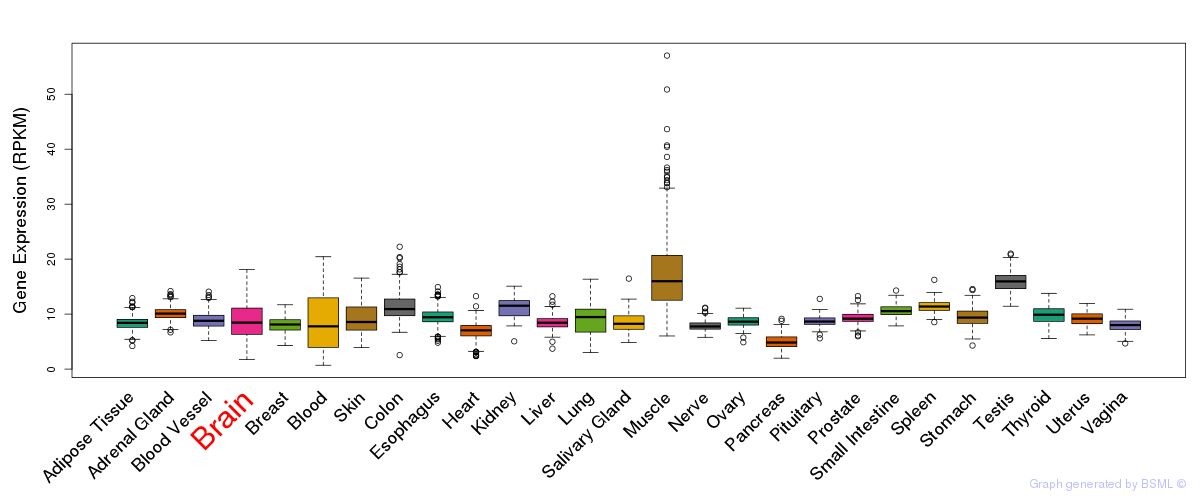
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
No co-expressed genes in brain regions
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| KEGG MTOR SIGNALING PATHWAY | 52 | 40 | All SZGR 2.0 genes in this pathway |
| KEGG INSULIN SIGNALING PATHWAY | 137 | 103 | All SZGR 2.0 genes in this pathway |
| REACTOME ANTIVIRAL MECHANISM BY IFN STIMULATED GENES | 66 | 45 | All SZGR 2.0 genes in this pathway |
| REACTOME INTERFERON SIGNALING | 159 | 116 | All SZGR 2.0 genes in this pathway |
| REACTOME IMMUNE SYSTEM | 933 | 616 | All SZGR 2.0 genes in this pathway |
| REACTOME CYTOKINE SIGNALING IN IMMUNE SYSTEM | 270 | 204 | All SZGR 2.0 genes in this pathway |
| LIU SOX4 TARGETS DN | 309 | 191 | All SZGR 2.0 genes in this pathway |
| ONKEN UVEAL MELANOMA UP | 783 | 507 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS DN | 431 | 263 | All SZGR 2.0 genes in this pathway |
| CASORELLI ACUTE PROMYELOCYTIC LEUKEMIA DN | 663 | 425 | All SZGR 2.0 genes in this pathway |
| DIAZ CHRONIC MEYLOGENOUS LEUKEMIA UP | 1382 | 904 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA DN | 320 | 184 | All SZGR 2.0 genes in this pathway |
| MULLIGHAN MLL SIGNATURE 2 UP | 418 | 263 | All SZGR 2.0 genes in this pathway |
| DODD NASOPHARYNGEAL CARCINOMA DN | 1375 | 806 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE KERATINOCYTE UP | 530 | 342 | All SZGR 2.0 genes in this pathway |
| VANHARANTA UTERINE FIBROID WITH 7Q DELETION DN | 36 | 23 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN DN | 1781 | 1082 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN DN | 770 | 415 | All SZGR 2.0 genes in this pathway |
| PUJANA BRCA1 PCC NETWORK | 1652 | 1023 | All SZGR 2.0 genes in this pathway |
| KOYAMA SEMA3B TARGETS DN | 411 | 249 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| LEI MYB TARGETS | 318 | 215 | All SZGR 2.0 genes in this pathway |
| ZAMORA NOS2 TARGETS UP | 69 | 41 | All SZGR 2.0 genes in this pathway |
| TSENG IRS1 TARGETS DN | 135 | 88 | All SZGR 2.0 genes in this pathway |
| RAMASWAMY METASTASIS UP | 66 | 43 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP | 566 | 371 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN | 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN | 1011 | 592 | All SZGR 2.0 genes in this pathway |
| GRADE COLON CANCER UP | 871 | 505 | All SZGR 2.0 genes in this pathway |
| GOLDRATH ANTIGEN RESPONSE | 346 | 192 | All SZGR 2.0 genes in this pathway |
| SHEDDEN LUNG CANCER POOR SURVIVAL A6 | 456 | 285 | All SZGR 2.0 genes in this pathway |
| YAGI AML WITH 11Q23 REARRANGED | 351 | 238 | All SZGR 2.0 genes in this pathway |
| CAIRO HEPATOBLASTOMA CLASSES UP | 605 | 377 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| YAO TEMPORAL RESPONSE TO PROGESTERONE CLUSTER 17 | 181 | 101 | All SZGR 2.0 genes in this pathway |
| JOHNSTONE PARVB TARGETS 3 DN | 918 | 550 | All SZGR 2.0 genes in this pathway |