| REACTOME HDL MEDIATED LIPID TRANSPORT
| 15 | 12 | All SZGR 2.0 genes in this pathway |
| REACTOME METABOLISM OF LIPIDS AND LIPOPROTEINS
| 478 | 302 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPID DIGESTION MOBILIZATION AND TRANSPORT
| 46 | 37 | All SZGR 2.0 genes in this pathway |
| REACTOME LIPOPROTEIN METABOLISM
| 28 | 24 | All SZGR 2.0 genes in this pathway |
| WATANABE RECTAL CANCER RADIOTHERAPY RESPONSIVE DN
| 92 | 51 | All SZGR 2.0 genes in this pathway |
| FULCHER INFLAMMATORY RESPONSE LECTIN VS LPS DN
| 463 | 290 | All SZGR 2.0 genes in this pathway |
| GARY CD5 TARGETS UP
| 473 | 314 | All SZGR 2.0 genes in this pathway |
| ELVIDGE HYPOXIA UP
| 171 | 112 | All SZGR 2.0 genes in this pathway |
| ENK UV RESPONSE EPIDERMIS DN
| 508 | 354 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN APOPTOSIS BY DOXORUBICIN UP
| 1142 | 669 | All SZGR 2.0 genes in this pathway |
| GRAESSMANN RESPONSE TO MC AND DOXORUBICIN UP
| 612 | 367 | All SZGR 2.0 genes in this pathway |
| WOOD EBV EBNA1 TARGETS UP
| 110 | 71 | All SZGR 2.0 genes in this pathway |
| SHETH LIVER CANCER VS TXNIP LOSS PAM1
| 229 | 137 | All SZGR 2.0 genes in this pathway |
| NUYTTEN EZH2 TARGETS UP
| 1037 | 673 | All SZGR 2.0 genes in this pathway |
| WEI MYCN TARGETS WITH E BOX
| 795 | 478 | All SZGR 2.0 genes in this pathway |
| BERTUCCI INVASIVE CARCINOMA DUCTAL VS LOBULAR UP
| 25 | 17 | All SZGR 2.0 genes in this pathway |
| MENSSEN MYC TARGETS
| 53 | 35 | All SZGR 2.0 genes in this pathway |
| SHEPARD BMYB MORPHOLINO UP
| 205 | 126 | All SZGR 2.0 genes in this pathway |
| LENAOUR DENDRITIC CELL MATURATION UP
| 114 | 84 | All SZGR 2.0 genes in this pathway |
| AFFAR YY1 TARGETS DN
| 234 | 137 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TROGLITAZONE UP
| 25 | 15 | All SZGR 2.0 genes in this pathway |
| RUAN RESPONSE TO TNF DN
| 84 | 50 | All SZGR 2.0 genes in this pathway |
| BLALOCK ALZHEIMERS DISEASE UP
| 1691 | 1088 | All SZGR 2.0 genes in this pathway |
| IVANOVA HEMATOPOIESIS INTERMEDIATE PROGENITOR
| 149 | 84 | All SZGR 2.0 genes in this pathway |
| WANG CISPLATIN RESPONSE AND XPC UP
| 202 | 115 | All SZGR 2.0 genes in this pathway |
| BURTON ADIPOGENESIS 6
| 189 | 112 | All SZGR 2.0 genes in this pathway |
| DOUGLAS BMI1 TARGETS UP
| 566 | 371 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR UP
| 953 | 554 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 6HR DN
| 911 | 527 | All SZGR 2.0 genes in this pathway |
| KRIGE RESPONSE TO TOSEDOSTAT 24HR DN
| 1011 | 592 | All SZGR 2.0 genes in this pathway |
| KONDO EZH2 TARGETS
| 245 | 148 | All SZGR 2.0 genes in this pathway |
| MASSARWEH TAMOXIFEN RESISTANCE DN
| 258 | 160 | All SZGR 2.0 genes in this pathway |
| MASSARWEH RESPONSE TO ESTRADIOL
| 61 | 47 | All SZGR 2.0 genes in this pathway |
| BOCHKIS FOXA2 TARGETS
| 425 | 261 | All SZGR 2.0 genes in this pathway |
| MATZUK OVULATION
| 14 | 10 | All SZGR 2.0 genes in this pathway |
| BOQUEST STEM CELL DN
| 216 | 143 | All SZGR 2.0 genes in this pathway |
| HUANG DASATINIB RESISTANCE DN
| 69 | 44 | All SZGR 2.0 genes in this pathway |
| BOYLAN MULTIPLE MYELOMA C D UP
| 139 | 95 | All SZGR 2.0 genes in this pathway |
| MILI PSEUDOPODIA HAPTOTAXIS DN
| 668 | 419 | All SZGR 2.0 genes in this pathway |
| MARTENS BOUND BY PML RARA FUSION
| 456 | 287 | All SZGR 2.0 genes in this pathway |
| DUTERTRE ESTRADIOL RESPONSE 6HR UP
| 229 | 149 | All SZGR 2.0 genes in this pathway |
| BHAT ESR1 TARGETS VIA AKT1 UP
| 281 | 183 | All SZGR 2.0 genes in this pathway |
| VANDESLUIS COMMD1 TARGETS GROUP 3 UP
| 89 | 50 | All SZGR 2.0 genes in this pathway |
| LEE BMP2 TARGETS UP
| 745 | 475 | All SZGR 2.0 genes in this pathway |
| FEVR CTNNB1 TARGETS DN
| 553 | 343 | All SZGR 2.0 genes in this pathway |
| GUO TARGETS OF IRS1 AND IRS2
| 98 | 67 | All SZGR 2.0 genes in this pathway |
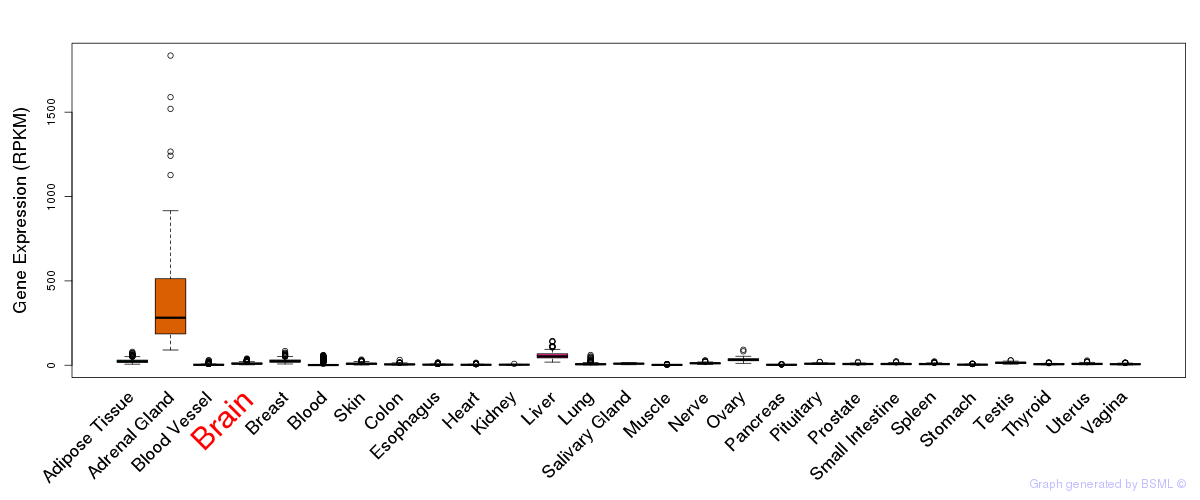
 Differentially methylated gene
Differentially methylated gene eQTL annotation
eQTL annotation