Gene Page: RPH3AL
Summary ?
| GeneID | 9501 |
| Symbol | RPH3AL |
| Synonyms | NOC2 |
| Description | rabphilin 3A-like (without C2 domains) |
| Reference | MIM:604881|HGNC:HGNC:10296|Ensembl:ENSG00000181031|HPRD:05345|Vega:OTTHUMG00000090273 |
| Gene type | protein-coding |
| Map location | 17p13.3 |
| Pascal p-value | 0.23 |
| Sherlock p-value | 0.15 |
| Fetal beta | -0.748 |
| eGene | Caudate basal ganglia Hypothalamus Nucleus accumbens basal ganglia Putamen basal ganglia Myers' cis & trans Meta |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:GWASdb | Genome-wide Association Studies | GWASdb records for schizophrenia | |
| CV:PGCnp | Genome-wide Association Study | GWAS |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs3923236 | chr1 | 201364433 | RPH3AL | 9501 | 0.19 | trans | ||
| rs62057049 | 17 | 65314 | RPH3AL | ENSG00000181031.11 | 1.563E-7 | 0 | 170731 | gtex_brain_putamen_basal |
| rs62057050 | 17 | 65610 | RPH3AL | ENSG00000181031.11 | 1.644E-7 | 0 | 170435 | gtex_brain_putamen_basal |
| rs7207457 | 17 | 65632 | RPH3AL | ENSG00000181031.11 | 2.247E-6 | 0 | 170413 | gtex_brain_putamen_basal |
| rs11868664 | 17 | 65720 | RPH3AL | ENSG00000181031.11 | 1.784E-7 | 0 | 170325 | gtex_brain_putamen_basal |
| rs72047060 | 17 | 66204 | RPH3AL | ENSG00000181031.11 | 9.798E-8 | 0 | 169841 | gtex_brain_putamen_basal |
| rs62057051 | 17 | 66563 | RPH3AL | ENSG00000181031.11 | 1.044E-7 | 0 | 169482 | gtex_brain_putamen_basal |
| rs8066107 | 17 | 66675 | RPH3AL | ENSG00000181031.11 | 5.8E-8 | 0 | 169370 | gtex_brain_putamen_basal |
| rs12941133 | 17 | 68686 | RPH3AL | ENSG00000181031.11 | 8.921E-8 | 0 | 167359 | gtex_brain_putamen_basal |
| rs12943724 | 17 | 71870 | RPH3AL | ENSG00000181031.11 | 8.087E-8 | 0 | 164175 | gtex_brain_putamen_basal |
| rs35292180 | 17 | 72161 | RPH3AL | ENSG00000181031.11 | 8.612E-8 | 0 | 163884 | gtex_brain_putamen_basal |
Section II. Transcriptome annotation
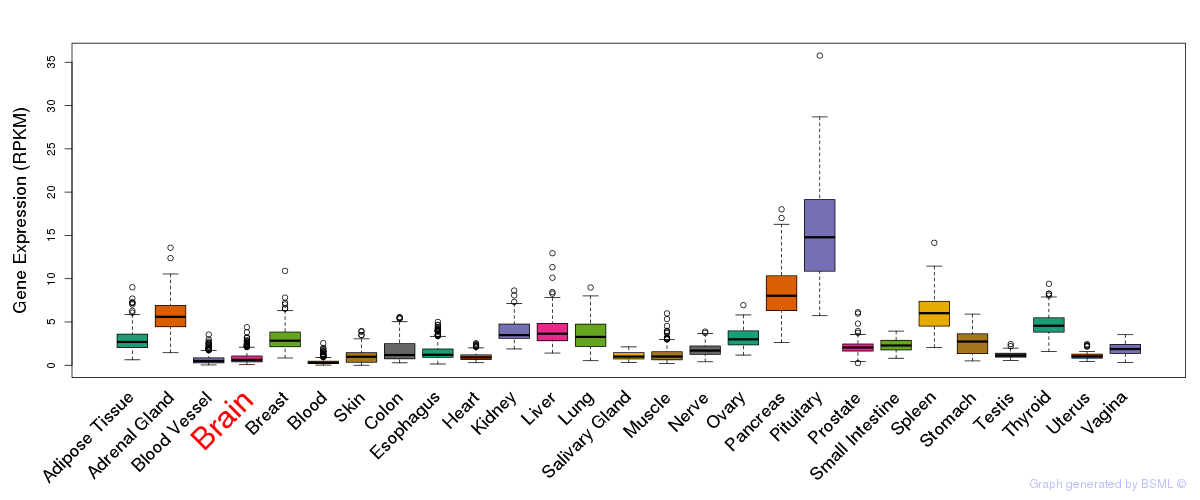
General gene expression (GTEx)
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MFSD4 | 0.96 | 0.91 |
| VIPR1 | 0.94 | 0.78 |
| DNM1 | 0.94 | 0.87 |
| CAMK2A | 0.93 | 0.84 |
| SLC17A7 | 0.93 | 0.92 |
| TYRO3 | 0.93 | 0.83 |
| CAMTA2 | 0.93 | 0.90 |
| KCNIP3 | 0.93 | 0.82 |
| AC105206.1 | 0.92 | 0.89 |
| PACSIN1 | 0.92 | 0.85 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| BCL7C | -0.58 | -0.67 |
| FADS2 | -0.52 | -0.43 |
| C9orf46 | -0.51 | -0.51 |
| TRAF4 | -0.50 | -0.50 |
| HEBP2 | -0.50 | -0.57 |
| GTF3C6 | -0.49 | -0.47 |
| DYNLT1 | -0.49 | -0.56 |
| TUBB2B | -0.49 | -0.47 |
| KIAA1949 | -0.49 | -0.35 |
| RPL23A | -0.49 | -0.52 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| RAB26 | V46133 | RAB26, member RAS oncogene family | Affinity Capture-Western | BioGRID | 12578829 |
| RAB27A | GS2 | HsT18676 | MGC117246 | RAB27 | RAM | RAB27A, member RAS oncogene family | - | HPRD,BioGRID | 12578829 |
| RAB27B | - | RAB27B, member RAS oncogene family | - | HPRD,BioGRID | 12578829 |
| RAB3A | - | RAB3A, member RAS oncogene family | - | HPRD,BioGRID | 11134008|12578829 |
| RAB3B | - | RAB3B, member RAS oncogene family | - | HPRD,BioGRID | 12578829 |
| RAB3C | - | RAB3C, member RAS oncogene family | Affinity Capture-Western | BioGRID | 12578829 |
| RAB3D | D2-2 | GOV | RAB16 | RAD3D | RAB3D, member RAS oncogene family | - | HPRD,BioGRID | 12578829 |
| RAB3GAP1 | DKFZp434A012 | KIAA0066 | P130 | RAB3GAP | RAB3GAP130 | WARBM1 | RAB3 GTPase activating protein subunit 1 (catalytic) | Reconstituted Complex | BioGRID | 14593078 |
| RAB8A | MEL | RAB8 | RAB8A, member RAS oncogene family | - | HPRD,BioGRID | 12578829 |
| RIMS2 | DKFZp781A0653 | KIAA0751 | OBOE | RAB3IP3 | RIM2 | regulating synaptic membrane exocytosis 2 | - | HPRD | 10707984 |
| UNC13B | MGC133279 | MGC133280 | MUNC13 | UNC13 | Unc13h2 | hmunc13 | unc-13 homolog B (C. elegans) | - | HPRD,BioGRID | 14593078 |
| ZYX | ESP-2 | HED-2 | zyxin | - | HPRD | 9367993 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| HOLLMANN APOPTOSIS VIA CD40 DN | 267 | 178 | All SZGR 2.0 genes in this pathway |
| DEURIG T CELL PROLYMPHOCYTIC LEUKEMIA UP | 368 | 234 | All SZGR 2.0 genes in this pathway |
| ODONNELL METASTASIS UP | 82 | 58 | All SZGR 2.0 genes in this pathway |
| CREIGHTON ENDOCRINE THERAPY RESISTANCE 4 | 307 | 185 | All SZGR 2.0 genes in this pathway |
| RIZKI TUMOR INVASIVENESS 3D UP | 210 | 124 | All SZGR 2.0 genes in this pathway |
| BILANGES SERUM SENSITIVE GENES | 90 | 54 | All SZGR 2.0 genes in this pathway |
| BILANGES RAPAMYCIN SENSITIVE VIA TSC1 AND TSC2 | 73 | 37 | All SZGR 2.0 genes in this pathway |