Gene Page: FXR2
Summary ?
| GeneID | 9513 |
| Symbol | FXR2 |
| Synonyms | FMR1L2|FXR2P |
| Description | FMR1 autosomal homolog 2 |
| Reference | MIM:605339|HGNC:HGNC:4024|Ensembl:ENSG00000129245|HPRD:05629|Vega:OTTHUMG00000178237 |
| Gene type | protein-coding |
| Map location | 17p13.1 |
| Pascal p-value | 0.468 |
| Sherlock p-value | 0.136 |
| Fetal beta | -0.439 |
| eGene | Myers' cis & trans |
| Support | Ascano FMRP targets |
Gene in Data Sources
| Gene set name | Method of gene set | Description | Info |
|---|---|---|---|
| CV:PGCnp | Genome-wide Association Study | GWAS | |
| PMID:cooccur | High-throughput literature-search | Systematic search in PubMed for genes co-occurring with SCZ keywords. A total of 3027 genes were included. | |
| Literature | High-throughput literature-search | Co-occurance with Schizophrenia keywords: schizophrenia,schizophrenias | Click to show details |
Section I. Genetics and epigenetics annotation
 eQTL annotation
eQTL annotation
| SNP ID | Chromosome | Position | eGene | Gene Entrez ID | pvalue | qvalue | TSS distance | eQTL type |
|---|---|---|---|---|---|---|---|---|
| rs10168927 | chr2 | 129503970 | FXR2 | 9513 | 0.19 | trans |
Section II. Transcriptome annotation
General gene expression (GTEx)
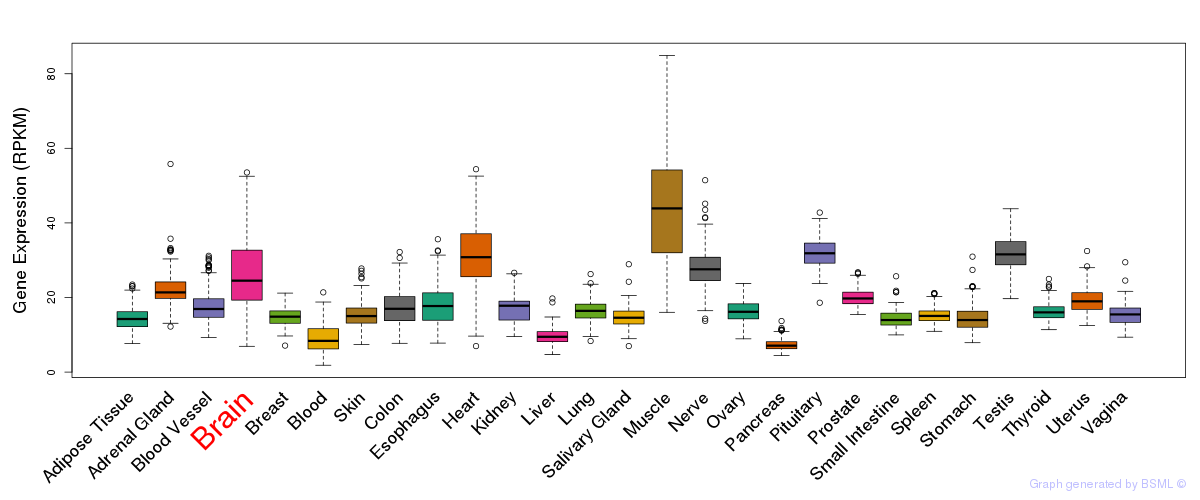
Gene expression during devlopment (BrainCloud)
Footnote:
A total of 269 time points ploted, with n=38 fetal samples (x=1:38). Each triangle represents one time point.
Gene expression of temporal and spatial changes (BrainSpan)
Footnote:
SC: sub-cortical regions; SM: sensory-motor regions; FC: frontal cortex; and TP: temporal-parietal cortex
ST1: fetal (13 - 26 postconception weeks), ST2: early infancy to late childhood (4 months to 11 years), and ST3: adolescence to adulthood (13 - 23 years)
The bar shown representes the lower 25% and upper 25% of the expression distribution.
Top co-expressed genes in brain regions
| Top 10 positively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| CDKN2AIP | 0.91 | 0.93 |
| TRA2B | 0.91 | 0.92 |
| SF3A3 | 0.91 | 0.91 |
| HNRNPK | 0.91 | 0.90 |
| TIAL1 | 0.90 | 0.91 |
| WDR33 | 0.90 | 0.90 |
| SUZ12 | 0.90 | 0.91 |
| DHX15 | 0.90 | 0.91 |
| ELP2 | 0.90 | 0.91 |
| H2AFY | 0.90 | 0.90 |
| Top 10 negatively co-expressed genes | ||
| Gene | Pearson's Correlation | Spearman's Correlation |
| MT-CO2 | -0.79 | -0.88 |
| AF347015.27 | -0.78 | -0.86 |
| AF347015.33 | -0.78 | -0.86 |
| AF347015.31 | -0.78 | -0.86 |
| MT-CYB | -0.78 | -0.87 |
| AF347015.8 | -0.78 | -0.87 |
| AF347015.15 | -0.76 | -0.86 |
| AF347015.2 | -0.74 | -0.85 |
| FXYD1 | -0.74 | -0.86 |
| AF347015.21 | -0.73 | -0.86 |
Section IV. Protein-protein interaction annotation
| Interactors | Aliases B | Official full name B | Experimental | Source | PubMed ID |
|---|---|---|---|---|---|
| AMOTL2 | LCCP | angiomotin like 2 | Two-hybrid | BioGRID | 16189514 |
| AP1M1 | AP47 | CLAPM2 | CLTNM | MU-1A | adaptor-related protein complex 1, mu 1 subunit | Two-hybrid | BioGRID | 16189514 |
| AP2M1 | AP50 | CLAPM1 | mu2 | adaptor-related protein complex 2, mu 1 subunit | Two-hybrid | BioGRID | 16189514 |
| ARL6IP1 | AIP1 | ARL6IP | ARMER | KIAA0069 | ADP-ribosylation factor-like 6 interacting protein 1 | Two-hybrid | BioGRID | 16189514 |
| C10orf62 | bA548K23.1 | chromosome 10 open reading frame 62 | Two-hybrid | BioGRID | 16189514 |
| C19orf50 | FLJ25480 | MGC2749 | MST096 | MSTP096 | chromosome 19 open reading frame 50 | Two-hybrid | BioGRID | 16189514 |
| C1orf149 | CDABP0189 | Eaf6 | FLJ11730 | NY-SAR-91 | chromosome 1 open reading frame 149 | Two-hybrid | BioGRID | 16189514 |
| CALCOCO2 | MGC17318 | NDP52 | calcium binding and coiled-coil domain 2 | Two-hybrid | BioGRID | 16189514 |
| CBS | HIP4 | cystathionine-beta-synthase | Two-hybrid | BioGRID | 16189514 |
| CCDC33 | FLJ23168 | FLJ32855 | MGC34145 | coiled-coil domain containing 33 | Two-hybrid | BioGRID | 16189514 |
| CCDC85B | DIPA | coiled-coil domain containing 85B | Two-hybrid | BioGRID | 16189514 |
| CYFIP1 | FLJ45151 | P140SRA-1 | SHYC | SRA1 | cytoplasmic FMR1 interacting protein 1 | Affinity Capture-Western Two-hybrid | BioGRID | 11438699 |
| DPPA2 | PESCRG1 | developmental pluripotency associated 2 | Two-hybrid | BioGRID | 16189514 |
| FBP1 | FBP | fructose-1,6-bisphosphatase 1 | Two-hybrid | BioGRID | 16189514 |
| FMR1 | FMRP | FRAXA | MGC87458 | POF | POF1 | fragile X mental retardation 1 | - | HPRD,BioGRID | 7489725 |8668200 |10567518 |
| FTH1 | FHC | FTH | FTHL6 | MGC104426 | PIG15 | PLIF | ferritin, heavy polypeptide 1 | Two-hybrid | BioGRID | 16189514 |
| FXR1 | - | fragile X mental retardation, autosomal homolog 1 | - | HPRD,BioGRID | 7489725 |8668200 |
| FXR2 | FMR1L2 | fragile X mental retardation, autosomal homolog 2 | - | HPRD,BioGRID | 7489725 |
| GKAP1 | FKSG21 | FLJ25469 | GKAP42 | G kinase anchoring protein 1 | Two-hybrid | BioGRID | 16189514 |
| HNRNPC | C1 | C2 | HNRNP | HNRPC | MGC104306 | MGC105117 | MGC117353 | MGC131677 | SNRPC | heterogeneous nuclear ribonucleoprotein C (C1/C2) | Two-hybrid | BioGRID | 16189514 |
| KCNRG | DLTET | potassium channel regulator | Two-hybrid | BioGRID | 16189514 |
| KCTD4 | bA321C24.3 | potassium channel tetramerisation domain containing 4 | Two-hybrid | BioGRID | 16189514 |
| KIAA1217 | DKFZp761L0424 | MGC31990 | SKT | KIAA1217 | Two-hybrid | BioGRID | 16189514 |
| KRT20 | CD20 | CK20 | K20 | KRT21 | MGC35423 | keratin 20 | Two-hybrid | BioGRID | 16189514 |
| LCMT1 | CGI-68 | LCMT | PPMT1 | leucine carboxyl methyltransferase 1 | Affinity Capture-Western Two-hybrid | BioGRID | 16189514 |
| LDOC1 | BCUR1 | Mar7 | Mart7 | leucine zipper, down-regulated in cancer 1 | Two-hybrid | BioGRID | 16189514 |
| MBIP | - | MAP3K12 binding inhibitory protein 1 | Two-hybrid | BioGRID | 16189514 |
| MCRS1 | ICP22BP | INO80Q | MCRS2 | MSP58 | P78 | microspherule protein 1 | Two-hybrid | BioGRID | 16189514 |
| MED7 | CRSP33 | CRSP9 | MGC12284 | mediator complex subunit 7 | Affinity Capture-MS | BioGRID | 17353931 |
| NCK2 | GRB4 | NCKbeta | NCK adaptor protein 2 | Two-hybrid | BioGRID | 16189514 |
| NECAB2 | EFCBP2 | N-terminal EF-hand calcium binding protein 2 | Two-hybrid | BioGRID | 16189514 |
| NONO | NMT55 | NRB54 | P54 | P54NRB | non-POU domain containing, octamer-binding | Two-hybrid | BioGRID | 16189514 |
| NT5C2 | GMP | NT5B | PNT5 | cN-II | 5'-nucleotidase, cytosolic II | Two-hybrid | BioGRID | 16189514 |
| PAICS | ADE2 | ADE2H1 | AIRC | DKFZp781N1372 | MGC1343 | MGC5024 | PAIS | phosphoribosylaminoimidazole carboxylase, phosphoribosylaminoimidazole succinocarboxamide synthetase | Two-hybrid | BioGRID | 16189514 |
| PCBD1 | DCOH | PCBD | PCD | PHS | pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha | Two-hybrid | BioGRID | 16189514 |
| PSME3 | Ki | PA28-gamma | PA28G | REG-GAMMA | proteasome (prosome, macropain) activator subunit 3 (PA28 gamma; Ki) | Two-hybrid | BioGRID | 16189514 |
| RABAC1 | PRA1 | PRAF1 | YIP3 | Rab acceptor 1 (prenylated) | Two-hybrid | BioGRID | 16189514 |
| RALYL | HNRPCL3 | RALY RNA binding protein-like | Two-hybrid | BioGRID | 16189514 |
| RBBP8 | CTIP | RIM | retinoblastoma binding protein 8 | Two-hybrid | BioGRID | 16189514 |
| RBMX | HNRPG | RBMXP1 | RBMXRT | RNMX | hnRNP-G | RNA binding motif protein, X-linked | Two-hybrid | BioGRID | 16189514 |
| RBPMS | HERMES | RNA binding protein with multiple splicing | Two-hybrid | BioGRID | 16189514 |
| RPIA | RPI | ribose 5-phosphate isomerase A | Two-hybrid | BioGRID | 16189514 |
| RTN3 | ASYIP | HAP | NSPL2 | NSPLII | RTN3-A1 | reticulon 3 | Two-hybrid | BioGRID | 16189514 |
| RTN4 | ASY | NI220/250 | NOGO | NOGO-A | NOGOC | NSP | NSP-CL | Nbla00271 | Nbla10545 | Nogo-B | Nogo-C | RTN-X | RTN4-A | RTN4-B1 | RTN4-B2 | RTN4-C | reticulon 4 | Two-hybrid | BioGRID | 16189514 |
| SNAP23 | HsT17016 | SNAP23A | SNAP23B | synaptosomal-associated protein, 23kDa | Two-hybrid | BioGRID | 16189514 |
| SSSCA1 | p27 | Sjogren syndrome/scleroderma autoantigen 1 | Two-hybrid | BioGRID | 16189514 |
| THAP1 | FLJ10477 | MGC33014 | THAP domain containing, apoptosis associated protein 1 | Two-hybrid | BioGRID | 16189514 |
| TNNT1 | ANM | FLJ98147 | MGC104241 | STNT | TNT | TNTS | troponin T type 1 (skeletal, slow) | Two-hybrid | BioGRID | 16189514 |
| TRAF2 | MGC:45012 | TRAP | TRAP3 | TNF receptor-associated factor 2 | Two-hybrid | BioGRID | 16189514 |
| TRIM29 | ATDC | FLJ36085 | tripartite motif-containing 29 | Two-hybrid | BioGRID | 16189514 |
| TRIM37 | KIAA0898 | MUL | POB1 | TEF3 | tripartite motif-containing 37 | Two-hybrid | BioGRID | 16189514 |
| TSC22D4 | THG-1 | THG1 | TSC22 domain family, member 4 | Two-hybrid | BioGRID | 16189514 |
| ZBTB8 | BOZF1 | FLJ90065 | MGC17919 | zinc finger and BTB domain containing 8 | Two-hybrid | BioGRID | 16189514 |
| ZNF451 | COASTER | FLJ23421 | FLJ53246 | FLJ90693 | KIAA0576 | KIAA1702 | MGC26701 | dJ417I1.1 | zinc finger protein 451 | Two-hybrid | BioGRID | 16189514 |
Section V. Pathway annotation
| Pathway name | Pathway size | # SZGR 2.0 genes in pathway | Info |
|---|---|---|---|
| DOANE RESPONSE TO ANDROGEN UP | 184 | 125 | All SZGR 2.0 genes in this pathway |
| GINESTIER BREAST CANCER ZNF217 AMPLIFIED DN | 335 | 193 | All SZGR 2.0 genes in this pathway |
| JAZAG TGFB1 SIGNALING VIA SMAD4 UP | 108 | 66 | All SZGR 2.0 genes in this pathway |
| BENPORATH MYC MAX TARGETS | 775 | 494 | All SZGR 2.0 genes in this pathway |
| AMIT EGF RESPONSE 480 HELA | 164 | 118 | All SZGR 2.0 genes in this pathway |
| HASLINGER B CLL WITH 17P13 DELETION | 21 | 15 | All SZGR 2.0 genes in this pathway |
| TAVOR CEBPA TARGETS DN | 30 | 21 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 20HR UP | 240 | 152 | All SZGR 2.0 genes in this pathway |
| BROWNE HCMV INFECTION 16HR UP | 225 | 139 | All SZGR 2.0 genes in this pathway |
| MOOTHA PGC | 420 | 269 | All SZGR 2.0 genes in this pathway |
| HOSHIDA LIVER CANCER SUBCLASS S3 | 266 | 180 | All SZGR 2.0 genes in this pathway |
| DANG BOUND BY MYC | 1103 | 714 | All SZGR 2.0 genes in this pathway |
| KIM ALL DISORDERS OLIGODENDROCYTE NUMBER CORR UP | 756 | 494 | All SZGR 2.0 genes in this pathway |